
The citric acid cycle (CAC) – also known as the TCA cycle or the Krebs cycle – is a series of chemical reactions to release stored energy through the oxidation of acetyl-CoA derived from carbohydrates, fats, and proteins. The Krebs cycle is used by organisms that respire to generate energy, either by anaerobic respiration or aerobic respiration. In addition, the cycle provides precursors of certain amino acids, as well as the reducing agent NADH, that are used in numerous other reactions. Its central importance to many biochemical pathways suggests that it was one of the earliest components of metabolism and may have originated abiogenically. Even though it is branded as a 'cycle', it is not necessary for metabolites to follow only one specific route; at least three alternative segments of the citric acid cycle have been recognized.
A dehydrogenase is an enzyme belonging to the group of oxidoreductases that oxidizes a substrate by reducing an electron acceptor, usually NAD+/NADP+ or a flavin coenzyme such as FAD or FMN. Like all catalysts, they catalyze reverse as well as forward reactions, and in some cases this has physiological significance: for example, alcohol dehydrogenase catalyzes the oxididation of ethanol to acetaldehyde in animals, but in yeast it catalyzes the production of ethanol from acetaldehyde.
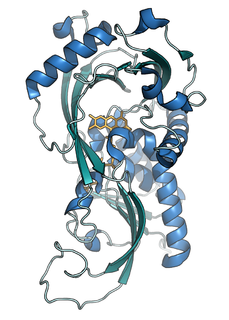
D-amino acid oxidase is an enzyme with the function on a molecular level to oxidize D-amino acids to the corresponding α-keto acids, producing ammonia and hydrogen peroxide. This results in a number of physiological effects in various systems, most notably the brain. The enzyme is most active toward neutral D-amino acids, and not active toward acidic D-amino acids. One of its most important targets in mammals is D-Serine in the central nervous system. By targeting this and other D-amino acids in vertebrates, DAAO is important in detoxification. The role in microorganisms is slightly different, breaking down D-amino acids to generate energy.
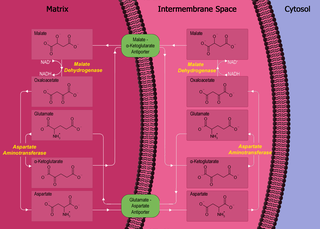
The malate-aspartate shuttle is a biochemical system for translocating electrons produced during glycolysis across the semipermeable inner membrane of the mitochondrion for oxidative phosphorylation in eukaryotes. These electrons enter the electron transport chain of the mitochondria via reduction equivalents to generate ATP. The shuttle system is required because the mitochondrial inner membrane is impermeable to NADH, the primary reducing equivalent of the electron transport chain. To circumvent this, malate carries the reducing equivalents across the membrane.
In enzymology, sarcosine dehydrogenase (EC 1.5.8.3) is a mitochondrial enzyme that catalyzes the chemical reaction N-demethylation of sarcosine to give glycine. This enzyme belongs to the family of oxidoreductases, specifically those acting on the CH-NH group of donor with other acceptors. The systematic name of this enzyme class is sarcosine:acceptor oxidoreductase (demethylating). Other names in common use include sarcosine N-demethylase, monomethylglycine dehydrogenase, and sarcosine:(acceptor) oxidoreductase (demethylating). Sarcosine dehydrogenase is closely related to dimethylglycine dehydrogenase, which catalyzes the demethylation reaction of dimethylglycine to sarcosine. Both sarcosine dehydrogenase and dimethylglycine dehydrogenase use FAD as a cofactor. Sarcosine dehydrogenase is linked by electron-transferring flavoprotein (ETF) to the respiratory redox chain. The general chemical reaction catalyzed by sarcosine dehydrogenase is:

In enzymology, a homoserine dehydrogenase (EC 1.1.1.3) is an enzyme that catalyzes the chemical reaction
In enzymology, a glycerol-3-phosphate dehydrogenase (NAD+) (EC 1.1.1.8) is an enzyme that catalyzes the chemical reaction
In enzymology, a gluconate 2-dehydrogenase (acceptor) is an enzyme that catalyzes the chemical reaction
In enzymology, a glycerol-3-phosphate oxidase (EC 1.1.3.21) is an enzyme that catalyzes the chemical reaction

In enzymology, an aspartate-semialdehyde dehydrogenase is an enzyme that is very important in the biosynthesis of amino acids in prokaryotes, fungi, and some higher plants. It forms an early branch point in the metabolic pathway forming lysine, methionine, leucine and isoleucine from aspartate. This pathway also produces diaminopimelate which plays an essential role in bacterial cell wall formation. There is particular interest in ASADH as disabling this enzyme proves fatal to the organism giving rise to the possibility of a new class of antibiotics, fungicides, and herbicides aimed at inhibiting it.
Aspartate dehydrogenase (EC 1.4.1.21) is an enzyme that catalyzes the chemical reaction
In enzymology, a D-aspartate oxidase (EC 1.4.3.1) is an enzyme that catalyzes the chemical reaction
In enzymology, a dimethylglycine dehydrogenase (EC 1.5.8.4) is an enzyme that catalyzes the chemical reaction

In enzymology, proline dehydrogenase (PRODH) is an enzyme of the oxidoreductase family, active in the oxidation of L-proline to (S)-1-pyrroline-5-carboxylate during proline catabolism. The end product of this reaction is then further oxidized in a (S)-1-pyrroline-5-carboxylate dehydrogenase (P5CDH)-dependent reaction of the proline metabolism, or spent to produce ornithine, a crucial metabolite of ornithine and arginine metabolism. The systematic name of this enzyme class is L-proline:quinone oxidoreductase. Other names in common use include L-proline dehydrogenase, L-proline oxidase,and L-proline:(acceptor) oxidoreductase. It employs one cofactor, FAD, which requires riboflavin.

FAD-dependent oxidoreductase domain-containing protein 1 (FOXRED1), also known as H17, or FP634 is an enzyme that in humans is encoded by the FOXRED1 gene. FOXRED1 is an oxidoreductase and complex I-specific molecular chaperone involved in the assembly and stabilization of NADH dehydrogenase (ubiquinone) also known as complex I, which is located in the mitochondrial inner membrane and is the largest of the five complexes of the electron transport chain. Mutations in FOXRED1 have been associated with Leigh syndrome and infantile-onset mitochondrial encephalopathy.
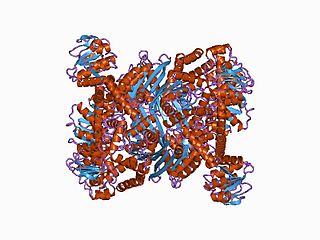
In molecular biology, the ELFV dehydrogenase family of enzymes include glutamate, leucine, phenylalanine and valine dehydrogenases. These enzymes are structurally and functionally related. They contain a Gly-rich region containing a conserved Lys residue, which has been implicated in the catalytic activity, in each case a reversible oxidative deamination reaction.

In molecular biology, the glucose-methanol-choline oxidoreductase family is a family of enzymes with oxidoreductase activity.
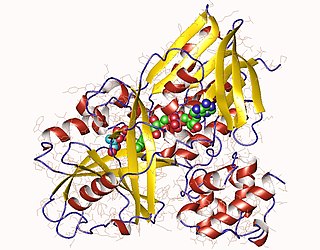
Glycerol-3-phosphate dehydrogenase (EC 1.1.5.3 is an enzyme with systematic name sn-glycerol 3-phosphate:quinone oxidoreductase. This enzyme catalyses the following chemical reaction
D-proline dehydrogenase is an enzyme with systematic name D-proline:acceptor oxidoreductase. This enzyme catalyses the following chemical reaction










