
The Bacillaceae-1 RNA motif is a conserved RNA structure identified by bioinformatics within bacteria in the family bacillaceae. The RNA is presumed to operate as a non-coding RNA, and is sometimes adjacent to operons containing ribosomal RNAs. The most characteristic feature is two terminal loops that have the nucleotide consensus RUCCU, where R is either A or G. The motif might be related to the Desulfotalea-1 RNA motif, as the motifs share some similarity in conserved features, and the Desulfotalea-1 RNA motif is also sometimes adjacent to ribosomal RNA operons.

The Bacteroid-trp RNA motif is a conserved RNA element detected by bioinformatics. It is found in the phylum Bacteroidota in the apparent 5' untranslated regions of genes that encode enzymes used in the synthesis of the amino acid tryptophan. A short open reading frame is found within the motif that encodes at least two tryptophan codons. Similar motifs have been identified regulating tryptophan genes in Pseudomonadota, but not in Bacteroidota. However, the Bacteroid-trp RNA motif likely operates via the same mechanism of attenuation.
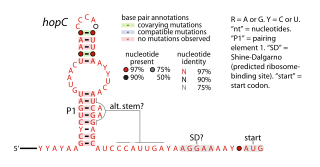
The hopC RNA motif is a predicted cis-regulatory element identified by a bioinformatic screen for conserved RNA secondary structures. hopC RNAs are exclusively found within bacteria classified within the genus Helicobacter, some of which are human pathogens that infect the stomach and can cause ulcers.

The JUMPstart RNA motif describes a conserved RNA-based secondary structure associated with JUMPstart elements. The 39-base-pair JUMPstart sequence describes a conserved element upstream of genes that participate in polysaccharide synthesis. The JUMPstart element has been shown to function as an RNA, and is present in the 5' untranslated regions of the genes it regulates.
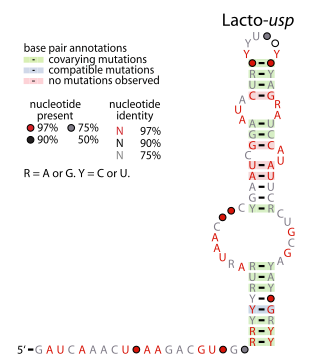
The Lacto-usp RNA motif is a conserved RNA structure identified in bacteria by bioinformatics. Lacto-usp RNAs are found exclusively in lactic acid bacteria, and exclusively in the possible 5′ untranslated regions of operons that contain a hypothetical gene and a usp gene. The usp gene encodes the universal stress protein. It was proposed that the Lacto-usp might correspond to the 6S RNA of the relevant species, because four of five of these species lack a predicted 6S RNA, and 6S RNAs commonly occur in 5′ UTRs of usp genes. However, given that the Lacto-usp RNA motif is much shorter than the standard 6S RNA structure, the function of Lacto-usp RNAs remains unclear.

The Methylobacterium-1 RNA motif is a conserved RNA structure discovered using bioinformatics. Almost all known examples of this RNA are found in DNA extracted from marine bacteria. However, one instance is predicted in Methylobacterium sp. 4-46, a species of alphaproteobacteria. The motif is presumed to function as a non-coding RNA.
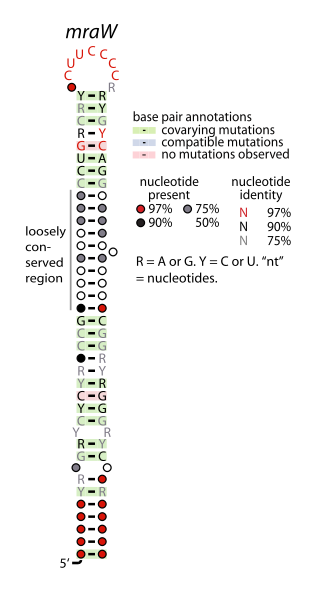
The mraW RNA motif is a conserved, structured RNA found in certain bacteria. Specifically, it is predicted in many, though not all, species of actinobacteria, and especially within the genus Mycobacterium. Structurally, the motif consists of a hairpin with a highly conserved terminal loop sequence. mraW RNAs are consistently in the presumed 5' untranslated regions of mraW genes. These mraW genes likely form operons with immediately downstream ftsI genes, and multiple types of mur genes. These genes are associated with peptidoglycan synthesis, and it was hypothesized that the mraW RNA motif might regulate these genes.
The nuoG RNA motif is a conserved RNA structure detected by bioinformatics. It is located in the presumed 5' untranslated regions of nuoG genes. This gene and the downstream genes probably comprise an operon that encodes various subunits of ubiquinone reductase enzyme.

PhotoRC RNA motifs refer to conserved RNA structures that are associated with genes acting in the photosynthetic reaction centre of photosynthetic bacteria. Two such RNA classes were identified and called the PhotoRC-I and PhotoRC-II motifs. PhotoRC-I RNAs were detected in the genomes of some cyanobacteria. Although no PhotoRC-II RNA has been detected in cyanobacteria, one is found in the genome of a purified phage that infects cyanobacteria. Both PhotoRC-I and PhotoRC-II RNAs are present in sequences derived from DNA that was extracted from uncultivated marine bacteria.

The psaA RNA motif describes a class of RNAs with a common secondary structure. psaA RNAs are exclusively found in locations that presumably correspond to the 5' untranslated regions of operons formed of psaA and psaB genes. For this reason, it was hypothesized that psaA RNAs function as cis-regulatory elements of these genes. The psaAB genes encode proteins that form subunits in the photosystem I structure used for photosynthesis. psaA RNAs have been detected only in cyanobacteria, which is consistent with their association with photosynthesis.
The Pseudomon-groES RNA motif is a conserved RNA structure identified in certain bacteria using bioinformatics. It is found in most species within the family Pseudomonadaceae, and is consistently located in the 5' untranslated regions of operons that contain groES genes. RNA transcripts of the groES genes in Pseudomonas aeruginosa where shown experimentally to be initiated at one of two start sites, from promoters called "P1" and "P2". The Pseudomon-groES RNA is in the 5' UTR of transcripts initiated from the P1 site, but is truncated in P2 transcripts. groES genes are involved in the cellular response to heat shock, but it is not thought that the Pseudomonas-groES RNA motif is involved in heat shock regulation. However, it is thought that the motif might regulate groES genes in response to other stimuli.

The Rhizobiales-2 RNA motif is a set of RNAs found in certain bacteria that are presumed to be homologous because they conserve a common primary and secondary structure. The motif was discovered using bioinformatics, and is found only within bacteria that belong to the order Hyphomicrobiales, in turn a kind of alphaproteobacteria. Because Rhizobiales-2 RNAs are not consistently located in proximity to genes of a consistent class or function, these RNAs are presumed to function as non-coding RNAs.

The SAM-Chlorobi RNA motif is a conserved RNA structure that was identified by bioinformatics. The RNAs are found only in bacteria classified as within the phylum Chlorobiota. These RNAs are always in the 5' untranslated regions of operons that contain metK and ahcY genes. metK genes encode methionine adenosyltransferase, which synthesizes S-adenosyl methionine (SAM), and ahcY genes encode S-adenosylhomocysteine hydrolase, which degrade the related metabolite S-Adenosyl-L-homocysteine (SAH). In fact all predicted metK and ahcY genes within Chlorobiota bacteria as of 2010 are preceded by predicted SAM-Chlorobi RNAs. Predicted promoter sequences are consistently found upstream of SAM-Chlorobi RNAs, and these promoter sequences imply that SAM-Chlorobi RNAs are indeed transcribed as RNAs. The promoter sequences are commonly associated with strong transcription in the phyla Chlorobiota and Bacteroidota, but are not used by most lineages of bacteria. The placement of SAM-Chlorobi RNAs suggests that they are involved in the regulation of the metK/ahcY operon through an unknown mechanism.

The sbcD RNA motif is a conserved RNA structure identified using bioinformatics. sbc RNAs are found some species of bacteria classified under the family Burkholderiaceae, and usually reside in plasmids. They are always located in what might be the 5' untranslated regions of operons that include sbcD genes. sbcD genes are involved in DNA repair.

The Termite-flg RNA motif is a conserved RNA structure identified by bioinformatics. Genomic sequences corresponding to Termite-flg RNAs have been identified only in uncultivated bacteria present in the termite hindgut. As of 2010 it has not been identified in the DNA of any cultivated species, and is thus an example of RNAs present in environmental samples.

The yjdF RNA motif is a conserved RNA structure identified using bioinformatics. Most yjdF RNAs are located in bacteria classified within the phylum Bacillota. A yjdF RNA is found in the presumed 5' untranslated region of the yjdF gene in Bacillus subtilis, and almost all yjdF RNAs are found in the 5' UTRs of homologs of this gene. The function of the yjdF gene is unknown, but the protein that it is predicted to encode is classified by the Pfam Database as DUF2992.
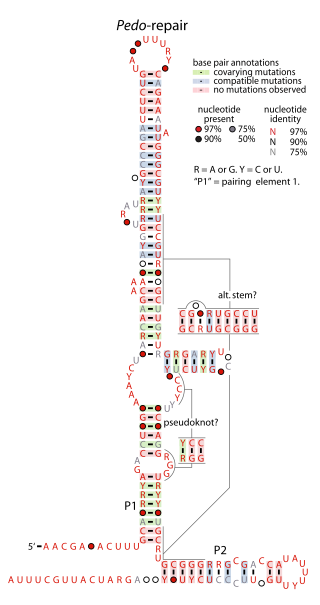
The Pedo-repair RNA motif is a conserved RNA structure identified by using bioinformatics. It has been detected in only one species of bacteria: Pedobacter sp. BAL39, within the phylum Bacteroidota. The motif might be in the 5′ untranslated regions of operons containing genes predicted to be involved in DNA repair or related to restriction enzymes.

The eps-Associated RNA element is a conserved RNA motif associated with exopolysaccharide (eps) or capsule biosynthesis genes in a subset of bacteria classified within the order Bacillales. It was initially discovered in Bacillus subtilis, located between the second and third gene in the eps operon. Deletion of the EAR element impairs biofilm formation.

The uup RNA motif is a conserved RNA structure that was discovered by bioinformatics. uup motif RNAs are found in Bacillota and Gammaproteobacteria.

The Zeta-pan RNA motif is a conserved RNA structure that was discovered by bioinformatics. Zeta-pan motif RNAs are found in Zetaproteobacteria.


















