
The Downstream-peptide motif refers to a conserved RNA structure identified by bioinformatics in the cyanobacterial genera Synechococcus and Prochlorococcus and one phage that infects such bacteria. It was also detected in marine samples of DNA from uncultivated bacteria, which are presumably other species of cyanobacteria.

The COG3943 RNA motif is a conserved RNA structure that was discovered by bioinformatics. COG3943 motifs are found in unknown bacteria whose genomic DNA was isolated from cow rumen. As of 2018, there is no specific, classified organism that is known to contain a COG3943 motif RNA.

The DUF2800 RNA motif is a conserved RNA structure that was discovered by bioinformatics. DUF2800 motif RNAs are found in Bacillota. DUF2800 RNAs are also predicted in the phyla Actinomycetota and Synergistota, although these RNAs are likely the result of recent horizontal gene transfer or conceivably sequence contamination.

The DUF805 RNA motif is a conserved RNA structure that was discovered by bioinformatics. The motif is subdivided into the DUF805 motif and the DUF805b motif, which have similar, but distinct secondary structures. Together, these motifs are found in Bacteroidota, Chlorobiota, and Pseudomonadota.
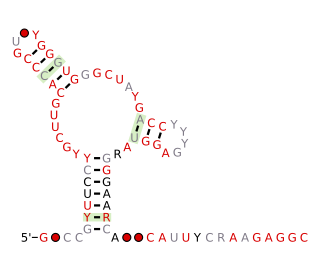
The Fibro-purF RNA motif is a conserved RNA structure that was discovered by bioinformatics. Fibro-purF motif RNAs are found in Fibrobacterota, a group of bacteria that are common in cow rumen. Additionally, the RNAs are found in metagenomic sequences of DNA isolated from cow rumen.

The Fibrobacter-1 RNA motif is a conserved RNA structure that was discovered by bioinformatics. Fibrobacter-1 motifs are found in organisms in the genus Fibrobacter, and in many metagenomic sequences.

The folE RNA motif, now known as the THF-II riboswitch, is a conserved RNA structure that was discovered by bioinformatics. folE motifs are found in Alphaproteobacteria.
The freshwater-2 RNA motif is a conserved RNA structure that was discovered by bioinformatics. Freshwater-2 motif RNAs are found in metagenomic sequences that are isolated from aquatic and especially freshwater environments. As of 2018, no freshwater-2 RNA has been identified in a classified organism.

The GA-cis RNA motif is a conserved RNA structure that was discovered by bioinformatics. GA-cis motif RNAs are found in one species classified within the phylum Bacillota: specifically, there are 9 predicted copies in Coprocuccus eutactus ATCC 27759.

The gntR-DTE RNA motif is a conserved RNA structure that was discovered by bioinformatics. gntR-DTE motifs are found in some, but not all species within the genus Streptomyces.
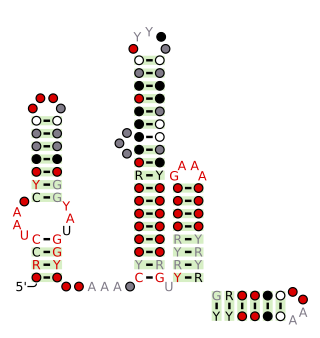
The Latescibacteria, OD1, OP11, TM7 RNA motif is a conserved RNA structure that was discovered by bioinformatics. LOOT motif RNAs are found in multiple bacterial phyla that have only recently been discovered, and are currently not well understood: Latescibacteria, OD1/Parcubacteria, OP11 AND TM7. In some cases, no specific organism has been isolated in the relevant phylum, but the existence of the bacterial phylum is known only through analysis of metagenomic sequences. Curiously, the LOOT motif is not known in any phylum that has been studied for a long time.
The Mahella-1 RNA motif is a conserved RNA structure that was discovered by bioinformatics. Seven Mahella-1 motif RNAs are found in Mahella australiensis, and no other organism has been observed to contain Mahella-1 RNAs.

The MDR-NUDIX RNA motif is a conserved RNA structure that was discovered by bioinformatics. The MDR-NUDIX motif is found in the poorly studied phylum TM7.

The nqrA-Marinomonas RNA motif is a conserved RNA structure that was discovered by bioinformatics. nqrA-Marinomonas motif RNAs are found in Marinomonas.
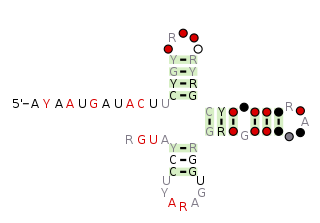
The pemK RNA motif is a conserved RNA structure that was discovered by bioinformatics. pemK motif RNAs are found in organisms within the phylum Bacillota, and is very widespread in this phylum.

The PGK RNA motif is a conserved RNA structure that was discovered by bioinformatics. PGK motif RNAs are found in metagenomic sequences isolated from the gastrointestinal tract of mammals. PGK RNAs have not yet been detected in a classified organism.

The Prevotella-2 RNA motif is a conserved RNA structure that was discovered by bioinformatics. Energetically stable tetraloops often occur in this motif. Prevotella-2 motif RNAs are found in the bacterial genus Prevotella.
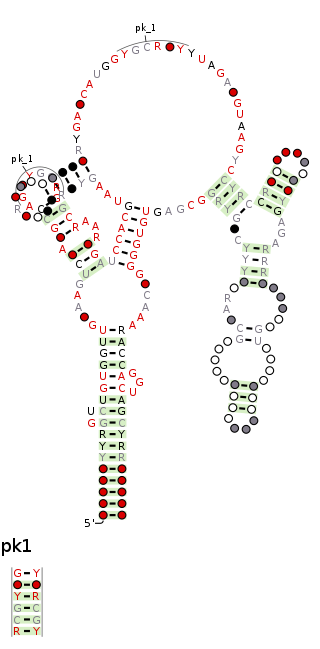
The raiA RNA motif is a conserved RNA structure that was discovered by bioinformatics. raiA motif RNAs are found in Actinomycetota and Bacillota, and have many conserved features—including conserved nucleotide positions, conserved secondary structures and associated protein-coding genes—in both of these phyla. Some conserved features of the raiA RNA motif suggest that they function as cis-regulatory elements, but other aspects of the motif suggest otherwise.

The uup RNA motif is a conserved RNA structure that was discovered by bioinformatics. uup motif RNAs are found in Bacillota and Gammaproteobacteria.

The xerDC RNA motif is a conserved RNA structure that was discovered by bioinformatics. xerDC motif RNAs are found in Clostridia.


















