
DNA ligase is a type of enzyme that facilitates the joining of DNA strands together by catalyzing the formation of a phosphodiester bond. It plays a role in repairing single-strand breaks in duplex DNA in living organisms, but some forms may specifically repair double-strand breaks. Single-strand breaks are repaired by DNA ligase using the complementary strand of the double helix as a template, with DNA ligase creating the final phosphodiester bond to fully repair the DNA.

A cloning vector is a small piece of DNA that can be stably maintained in an organism, and into which a foreign DNA fragment can be inserted for cloning purposes. The cloning vector may be DNA taken from a virus, the cell of a higher organism, or it may be the plasmid of a bacterium. The vector contains features that allow for the convenient insertion of a DNA fragment into the vector or its removal from the vector, for example through the presence of restriction sites. The vector and the foreign DNA may be treated with a restriction enzyme that cuts the DNA, and DNA fragments thus generated contain either blunt ends or overhangs known as sticky ends, and vector DNA and foreign DNA with compatible ends can then be joined by molecular ligation. After a DNA fragment has been cloned into a cloning vector, it may be further subcloned into another vector designed for more specific use.

In molecular biology, a library is a collection of genetic material fragments that are stored and propagated in a population of microbes through the process of molecular cloning. There are different types of DNA libraries, including cDNA libraries, genomic libraries and randomized mutant libraries. DNA library technology is a mainstay of current molecular biology, genetic engineering, and protein engineering, and the applications of these libraries depend on the source of the original DNA fragments. There are differences in the cloning vectors and techniques used in library preparation, but in general each DNA fragment is uniquely inserted into a cloning vector and the pool of recombinant DNA molecules is then transferred into a population of bacteria or yeast such that each organism contains on average one construct. As the population of organisms is grown in culture, the DNA molecules contained within them are copied and propagated.
A restriction digest is a procedure used in molecular biology to prepare DNA for analysis or other processing. It is sometimes termed DNA fragmentation, though this term is used for other procedures as well. In a restriction digest, DNA molecules are cleaved at specific restriction sites of 4-12 nucleotides in length by use of restriction enzymes which recognize these sequences.
DNA fragmentation is the separation or breaking of DNA strands into pieces. It can be done intentionally by laboratory personnel or by cells, or can occur spontaneously. Spontaneous or accidental DNA fragmentation is fragmentation that gradually accumulates in a cell. It can be measured by e.g. the Comet assay or by the TUNEL assay.

In molecular biology, subcloning is a technique used to move a particular DNA sequence from a parent vector to a destination vector.
Tandemly arrayed genes (TAGs) are a gene cluster created by tandem duplications, a process in which one gene is duplicated and the copy is found adjacent to the original. They serve to encode large numbers of genes at a time.

Karyolysis, and λύσις lysis from λύειν lyein, "to separate") is the complete dissolution of the chromatin of a dying cell due to the enzymatic degradation by endonucleases. The whole cell will eventually stain uniformly with eosin after karyolysis. It is usually associated with karyorrhexis and occurs mainly as a result of necrosis, while in apoptosis after karyorrhexis the nucleus usually dissolves into apoptotic bodies.

DNA laddering is a feature that can be observed when DNA fragments, resulting from Apoptosis DNA fragmentation are visualized after separation by gel electrophoresis the first described in 1980 by Andrew Wyllie at the University Edinburgh medical school DNA fragments can also be detected in cells that underwent necrosis, but when these DNA fragments after separation are subjected to gel electrophoresis no clear "ladder" pattern is apparent.

DNA shuffling, also known as molecular breeding, is an in vitro random recombination method to generate mutant genes for directed evolution and to enable a rapid increase in DNA library size. Three procedures for accomplishing DNA shuffling are molecular breeding which relies on homologous recombination or the similarity of the DNA sequences, restriction enzymes which rely on common restriction sites, and nonhomologous random recombination which requires the use of hairpins. In all of these techniques, the parent genes are fragmented and then recombined.
A genomic library is a collection of overlapping DNA fragments that together make up the total genomic DNA of a single organism. The DNA is stored in a population of identical vectors, each containing a different insert of DNA. In order to construct a genomic library, the organism's DNA is extracted from cells and then digested with a restriction enzyme to cut the DNA into fragments of a specific size. The fragments are then inserted into the vector using DNA ligase. Next, the vector DNA can be taken up by a host organism - commonly a population of Escherichia coli or yeast - with each cell containing only one vector molecule. Using a host cell to carry the vector allows for easy amplification and retrieval of specific clones from the library for analysis.
Cellular waste products are formed as a by-product of cellular respiration, a series of processes and reactions that generate energy for the cell, in the form of ATP. One example of cellular respiration creating cellular waste products are aerobic respiration and anaerobic respiration.

Apoptotic DNA fragmentation is a key feature of apoptosis, a type of programmed cell death. Apoptosis is characterized by the activation of endogenous endonucleases, particularly the caspase-3 activated DNase (CAD), with subsequent cleavage of nuclear DNA into internucleosomal fragments of roughly 180 base pairs (bp) and multiples thereof (360, 540 etc.). The apoptotic DNA fragmentation is being used as a marker of apoptosis and for identification of apoptotic cells either via the DNA laddering assay, the TUNEL assay, or the by detection of cells with fractional DNA content ("sub G1 cells") on DNA content frequency histograms e.g. as in the Nicoletti assay.
Topoisomerase-based cloning is a molecular biology technique in which DNA fragments are cloned into specific vectors without the requirement for DNA ligases. Taq polymerase has a nontemplate-dependent terminal transferase activity that adds a single deoxyadenosine (A) to the 3'-end of the PCR products. This characteristic is exploited in "sticky end" TOPO TA cloning. For "blunt end" TOPO cloning, the recipient vector does not have overhangs and blunt-ended DNA fragments can be cloned.

Caspase-activated DNase (CAD) or DNA fragmentation factor subunit beta is a protein that in humans is encoded by the DFFB gene. It breaks up the DNA during apoptosis and promotes cell differentiation. It is usually an inactive monomer inhibited by ICAD. This is cleaved before dimerization.
Gibson assembly is a molecular cloning method that allows for the joining of multiple DNA fragments in a single, isothermal reaction. It is named after its creator, Daniel G. Gibson, who is the chief technology officer and co-founder of the synthetic biology company, Telesis Bio. The technology is more efficient than manual plasmid genetic recombination methods, but remains expensive as it is still under patent.

Molecular cloning is a set of experimental methods in molecular biology that are used to assemble recombinant DNA molecules and to direct their replication within host organisms. The use of the word cloning refers to the fact that the method involves the replication of one molecule to produce a population of cells with identical DNA molecules. Molecular cloning generally uses DNA sequences from two different organisms: the species that is the source of the DNA to be cloned, and the species that will serve as the living host for replication of the recombinant DNA. Molecular cloning methods are central to many contemporary areas of modern biology and medicine.
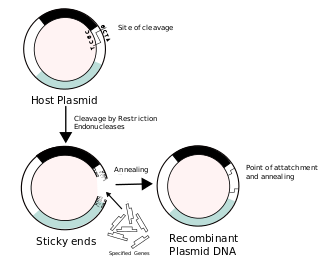
Recombinant DNA (rDNA), or molecular cloning, is the process by which a single gene, or segment of DNA, is isolated and amplified. Recombinant DNA is also known as in vitro recombination. A cloning vector is a DNA molecule that carries foreign DNA into a host cell, where it replicates, producing many copies of itself along with the foreign DNA. There are many types of cloning vectors such as plasmids and phages. In order to carry out recombination between vector and the foreign DNA, it is necessary the vector and DNA to be cloned by digestion, ligase the foreign DNA into the vector with the enzyme DNA ligase. And DNA is inserted by introducing the DNA into bacteria cells by transformation.

Ligation is the joining of two nucleotides, or two nucleic acid fragments, into a single polymeric chain through the action of an enzyme known as a ligase. The reaction involves the formation of a phosphodiester bond between the 3'-hydroxyl terminus of one nucleotide and the 5'-phosphoryl terminus of another nucleotide, which results in the two nucleotides being linked consecutively on a single strand. Ligation works in fundamentally the same way for both DNA and RNA. A cofactor is generally involved in the reaction, usually ATP or NAD+. Eukaryotic ligases belong to the ATP type, while the NAD+ type are found in bacteria (e.g. E. coli).
Rate-zonal centrifugation is a centrifugation technique employed to effectively separate particles of different sizes. The tube is first filled with different concentrations of sucrose or another solute establishing layers with different densities and viscosities, forming a density gradient, within which the particles to be separated are added. The larger particles will be able to travel to the bottom layer because they are more massive. The greater mass allows the particles to travel through layers with a greater viscosity, while the smaller particles will remain at the top, as they lack the mass to travel through the more viscous layers. Once the centrifugation is over, fractions are collected.














