
Enterobacteria phage λ is a bacterial virus, or bacteriophage, that infects the bacterial species Escherichia coli. It was discovered by Esther Lederberg in 1950. The wild type of this virus has a temperate life cycle that allows it to either reside within the genome of its host through lysogeny or enter into a lytic phase, during which it kills and lyses the cell to produce offspring. Lambda strains, mutated at specific sites, are unable to lysogenize cells; instead, they grow and enter the lytic cycle after superinfecting an already lysogenized cell.
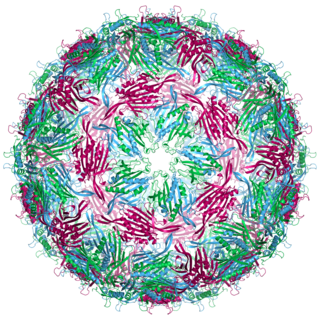
Bacteriophage MS2, commonly called MS2, is an icosahedral, positive-sense single-stranded RNA virus that infects the bacterium Escherichia coli and other members of the Enterobacteriaceae. MS2 is a member of a family of closely related bacterial viruses that includes bacteriophage f2, bacteriophage Qβ, R17, and GA.
Antitermination is the prokaryotic cell's aid to fix premature termination of RNA synthesis during the transcription of RNA. It occurs when the RNA polymerase ignores the termination signal and continues elongating its transcript until a second signal is reached. Antitermination provides a mechanism whereby one or more genes at the end of an operon can be switched either on or off, depending on the polymerase either recognizing or not recognizing the termination signal.

fis is an E. coli gene encoding the Fis protein. The regulation of this gene is more complex than most other genes in the E. coli genome, as Fis is an important protein which regulates expression of other genes. It is supposed that fis is regulated by H-NS, IHF and CRP. It also regulates its own expression (autoregulation). Fis is one of the most abundant DNA binding proteins in Escherichia coli under nutrient-rich growth conditions.

The Ykok leader or M-box is a Mg2+-sensing RNA structure that controls the expression of Magnesium ion transport proteins in bacteria. It is a distinct structure to the Magnesium responsive RNA element.

The c4 antisense RNA is a non-coding RNA used by certain phages that infect bacteria. It was initially identified in the P1 and P7 phages of E. coli. The identification of c4 antisense RNAs solved the mystery of the mechanism for regulation of the ant gene, which is an anti-repressor.

The Downstream-peptide motif refers to a conserved RNA structure identified by bioinformatics in the cyanobacterial genera Synechococcus and Prochlorococcus and one phage that infects such bacteria. It was also detected in marine samples of DNA from uncultivated bacteria, which are presumably other species of cyanobacteria.
The wcaG RNA motif is an RNA structure conserved in some bacteria that was detected by bioinformatics. wcaG RNAs are found in certain phages that infect cyanobacteria. Most known wcaG RNAs were found in sequences of DNA extracted from uncultivated marine bacteria. wcaG RNAs might function as cis-regulatory elements, in view of their consistent location in the possible 5' untranslated regions of genes. It was suggested the wcaG RNAs might further function as riboswitches.

PhotoRC RNA motifs refer to conserved RNA structures that are associated with genes acting in the photosynthetic reaction centre of photosynthetic bacteria. Two such RNA classes were identified and called the PhotoRC-I and PhotoRC-II motifs. PhotoRC-I RNAs were detected in the genomes of some cyanobacteria. Although no PhotoRC-II RNA has been detected in cyanobacteria, one is found in the genome of a purified phage that infects cyanobacteria. Both PhotoRC-I and PhotoRC-II RNAs are present in sequences derived from DNA that was extracted from uncultivated marine bacteria.

The D12-methyl RNA motif is a conserved RNA structure that was discovered by bioinformatics. D12-methyl motifs are found in metagenomic DNA samples, and have not yet been found in a classified organism.

The DUF2693 RNA motif is a conserved RNA structure that was discovered by bioinformatics. DUF2693 motif RNAs are found in Porphyromonas.

The DUF2800 RNA motif is a conserved RNA structure that was discovered by bioinformatics. DUF2800 motif RNAs are found in Bacillota. DUF2800 RNAs are also predicted in the phyla Actinomycetota and Synergistota, although these RNAs are likely the result of recent horizontal gene transfer or conceivably sequence contamination.

The DUF2815 RNA motif is a conserved RNA structure that was discovered by bioinformatics. As of 2018, the DUF2815 motif has not been identified in any classified organism, but is known through metagenomic sequences isolated from environmental sources.
The freshwater-2 RNA motif is a conserved RNA structure that was discovered by bioinformatics. Freshwater-2 motif RNAs are found in metagenomic sequences that are isolated from aquatic and especially freshwater environments. As of 2018, no freshwater-2 RNA has been identified in a classified organism.
malK RNA motifs are conserved RNA structures that were discovered by bioinformatics. They are defined by being consistently located upstream of malK genes, which encode an ATPase that is used by transporters whose ligand is likely a kind of sugar. Most of these genes are annotated either as transporting maltose or glycerol-3-phosphate, however the substrate of the transporters associated with malK motif RNAs has not been experimentally determined. All known types of malK RNA motif are generally located nearby to the Shine-Dalgarno sequence of the downstream gene.
The mcrA RNA motif is a conserved RNA structure that was discovered by bioinformatics.

The Mu-like gpT Downstream Element RNA motif is a conserved RNA structure that was discovered by bioinformatics. The Mu-gpT-DE motif is only found in metagenomic sequences arising from unknown organisms.

The Proteo-phage-1 RNA motif is a conserved RNA structure that was discovered by bioinformatics. Energetically stable tetraloops often occur in this motif.
The sbcC RNA motif is a conserved RNA structure that was discovered by bioinformatics. The sbcC motif has, as of 2018, only been detected in metagenomic sequences, and the identities of organisms that contain these RNAs is unknown.

The ssNA-helicase RNA motif is a conserved RNA structure that was discovered by bioinformatics. Although the ssNA-helicase motif was published as an RNA candidate, there is some reason to suspect that it might function as a single-stranded DNA. In terms of secondary structure, RNA and DNA are difficult to distinguish when only sequence information is available.














