Related Research Articles

Proteomics is the large-scale study of proteins. Proteins are vital parts of living organisms, with many functions such as the formation of structural fibers of muscle tissue, enzymatic digestion of food, or synthesis and replication of DNA. In addition, other kinds of proteins include antibodies that protect an organism from infection, and hormones that send important signals throughout the body.
Glycomics is the comprehensive study of glycomes, including genetic, physiologic, pathologic, and other aspects. Glycomics "is the systematic study of all glycan structures of a given cell type or organism" and is a subset of glycobiology. The term glycomics is derived from the chemical prefix for sweetness or a sugar, "glyco-", and was formed to follow the omics naming convention established by genomics and proteomics.

In molecular biology, post-translational modification (PTM) is the covalent process of changing proteins following protein biosynthesis. PTMs may involve enzymes or occur spontaneously. Proteins are created by ribosomes, which translate mRNA into polypeptide chains, which may then change to form the mature protein product. PTMs are important components in cell signalling, as for example when prohormones are converted to hormones.

Glycoproteins are proteins which contain oligosaccharide chains covalently attached to amino acid side-chains. The carbohydrate is attached to the protein in a cotranslational or posttranslational modification. This process is known as glycosylation. Secreted extracellular proteins are often glycosylated.
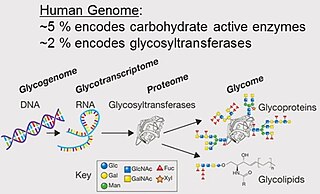
A glycome is the entire complement or complete set of all sugars, whether free or chemically bound in more complex molecules, of an organism. An alternative definition is the entirety of carbohydrates in a cell. The glycome may in fact be one of the most complex entities in nature. "Glycomics, analogous to genomics and proteomics, is the systematic study of all glycan structures of a given cell type or organism" and is a subset of glycobiology.
Defined in the narrowest sense, glycobiology is the study of the structure, biosynthesis, and biology of saccharides that are widely distributed in nature. Sugars or saccharides are essential components of all living things and aspects of the various roles they play in biology are researched in various medical, biochemical and biotechnological fields.
Glycosylation is the reaction in which a carbohydrate, i.e. a glycosyl donor, is attached to a hydroxyl or other functional group of another molecule in order to form a glycoconjugate. In biology, glycosylation usually refers to an enzyme-catalysed reaction, whereas glycation may refer to a non-enzymatic reaction.

A protein isoform, or "protein variant", is a member of a set of highly similar proteins that originate from a single gene or gene family and are the result of genetic differences. While many perform the same or similar biological roles, some isoforms have unique functions. A set of protein isoforms may be formed from alternative splicings, variable promoter usage, or other post-transcriptional modifications of a single gene; post-translational modifications are generally not considered. Through RNA splicing mechanisms, mRNA has the ability to select different protein-coding segments (exons) of a gene, or even different parts of exons from RNA to form different mRNA sequences. Each unique sequence produces a specific form of a protein.
The terms glycans and polysaccharides are defined by IUPAC as synonyms meaning "compounds consisting of a large number of monosaccharides linked glycosidically". However, in practice the term glycan may also be used to refer to the carbohydrate portion of a glycoconjugate, such as a glycoprotein, glycolipid, or a proteoglycan, even if the carbohydrate is only an oligosaccharide. Glycans usually consist solely of O-glycosidic linkages of monosaccharides. For example, cellulose is a glycan composed of β-1,4-linked D-glucose, and chitin is a glycan composed of β-1,4-linked N-acetyl-D-glucosamine. Glycans can be homo- or heteropolymers of monosaccharide residues, and can be linear or branched.
Biomarker discovery is a medical term describing the process by which biomarkers are discovered. Many commonly used blood tests in medicine are biomarkers. There is interest in biomarker discovery on the part of the pharmaceutical industry; blood-test or other biomarkers could serve as intermediate markers of disease in clinical trials, and as possible drug targets.

Chi-Huey Wong is a Taiwanese-American biochemist. He is currently the Scripps Family Chair Professor at the Scripps Research Institute, California in the department of chemistry. He is a member of the United States National Academy of Sciences, as awarded the 2014 Wolf Prize in Chemistry and 2015 RSC Robert Robinson Award. Wong is also the holder of more than 100 patents and publisher of 700 more scholarly academic research papers under his name.
Surface-enhanced laser desorption/ionization (SELDI) is a soft ionization method in mass spectrometry (MS) used for the analysis of protein mixtures. It is a variation of matrix-assisted laser desorption/ionization (MALDI). In MALDI, the sample is mixed with a matrix material and applied to a metal plate before irradiation by a laser, whereas in SELDI, proteins of interest in a sample become bound to a surface before MS analysis. The sample surface is a key component in the purification, desorption, and ionization of the sample. SELDI is typically used with time-of-flight (TOF) mass spectrometers and is used to detect proteins in tissue samples, blood, urine, or other clinical samples, however, SELDI technology can potentially be used in any application by simply modifying the sample surface.

Protein mass spectrometry refers to the application of mass spectrometry to the study of proteins. Mass spectrometry is an important method for the accurate mass determination and characterization of proteins, and a variety of methods and instrumentations have been developed for its many uses. Its applications include the identification of proteins and their post-translational modifications, the elucidation of protein complexes, their subunits and functional interactions, as well as the global measurement of proteins in proteomics. It can also be used to localize proteins to the various organelles, and determine the interactions between different proteins as well as with membrane lipids.
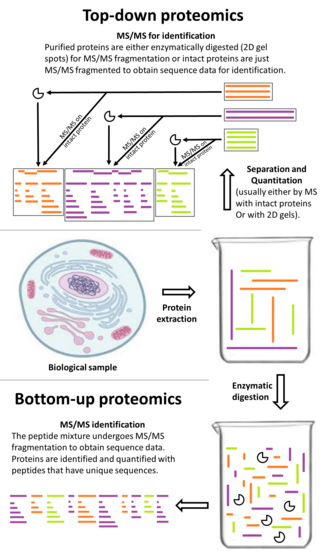
Top-down proteomics is a method of protein identification that either uses an ion trapping mass spectrometer to store an isolated protein ion for mass measurement and tandem mass spectrometry (MS/MS) analysis or other protein purification methods such as two-dimensional gel electrophoresis in conjunction with MS/MS. Top-down proteomics is capable of identifying and quantitating unique proteoforms through the analysis of intact proteins. The name is derived from the similar approach to DNA sequencing. During mass spectrometry intact proteins are typically ionized by electrospray ionization and trapped in a Fourier transform ion cyclotron resonance, quadrupole ion trap or Orbitrap mass spectrometer. Fragmentation for tandem mass spectrometry is accomplished by electron-capture dissociation or electron-transfer dissociation. Effective fractionation is critical for sample handling before mass-spectrometry-based proteomics. Proteome analysis routinely involves digesting intact proteins followed by inferred protein identification using mass spectrometry (MS). Top-down MS (non-gel) proteomics interrogates protein structure through measurement of an intact mass followed by direct ion dissociation in the gas phase.
Anne Dell is an Australian biochemist specialising in the study of glycomics and the carbohydrate structures that modify proteins. Anne's work could be used to figure out how pathogens such as HIV are able to evade termination by the immune system which could be applied toward understanding how this occurs in fetuses. Her research has also led to the development of higher sensitivity mass spectroscopy techniques which have allowed for the better studying of the structure of carbohydrates. Anne also established GlycoTRIC at Imperial College London, a research center that allows for glycobiology to be better understood in biomedical applications. She is currently Professor of Carbohydrate Biochemistry and Head of the Department of Life Sciences at Imperial College London. Dell's other contributions to the study of Glycobiology are the additions she has made to the textbook "Essentials of Glycobiology" Dell was appointed Commander of the Order of the British Empire (CBE) in the 2009 Birthday Honours.
The eastern blot, or eastern blotting, is a biochemical technique used to analyze protein post-translational modifications including the addition of lipids, phosphates, and glycoconjugates. It is most often used to detect carbohydrate epitopes. Thus, eastern blot can be considered an extension of the biochemical technique of western blot. Multiple techniques have been described by the term "eastern blot(ting)", most use phosphoprotein blotted from sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gel on to a polyvinylidene fluoride or nitrocellulose membrane. Transferred proteins are analyzed for post-translational modifications using probes that may detect lipids, carbohydrate, phosphorylation or any other protein modification. Eastern blotting should be used to refer to methods that detect their targets through specific interaction of the post-translational modifications and the probe, distinguishing them from a standard far-western blot. In principle, eastern blotting is similar to lectin blotting.
Hui Zhang is a professor of Pathology at Johns Hopkins University. She specializes in analysis of glycoproteins and other protein modifications on the proteome scale. Her most cited article is Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry.

Albert J.R. Heck is a Dutch scientist and professor at Utrecht University, the Netherlands in the field of mass spectrometry and proteomics. He is known for his work on technologies to study proteins in their natural environment, with the aim to understand their biological function. Albert Heck was awarded the Spinoza Prize in 2017, the highest scientific award in the Netherlands.
Translational glycobiology or applied glycobiology is the branch of glycobiology and glycochemistry that focuses on developing new pharmaceuticals through glycomics and glycoengineering. Although research in this field presents many difficulties, translational glycobiology presents applications with therapeutic glycoconjugates, with treating various bone diseases, and developing therapeutic cancer vaccines and other targeted therapies. Some mechanisms of action include using the glycan for drug targeting, engineering protein glycosylation for better efficacy, and glycans as drugs themselves.
Nicki Packer FRSC is an Australian college professor and researcher. She currently serves as a distinguished professor of glycoproteomics in the School of Natural Sciences at Macquarie University and principal research leader at Griffith University's Institute for Glycomics. Packer is a Fellow of the Royal Society of Chemistry and in 2021 received the Distinguished Achievement in Proteomic Sciences Award from the Human Proteome Organization. Her research focuses on biological functional of glycoconjugates by linking glycomics with proteomics and bioinformatics.
References
- ↑ Tissot B, North SJ, Ceroni A, Pang PC, Panico M, Rosati F, et al. (June 2009). "Glycoproteomics: past, present and future". FEBS Letters. 583 (11): 1728–1735. doi:10.1016/j.febslet.2009.03.049. PMC 2753369 . PMID 19328791.
- 1 2 Doerr A (January 2012). "Glycoproteomics". Nature Methods. 9 (1): 36. doi: 10.1038/nmeth.1821 . ISSN 1548-7091.
- ↑ Pan S, Chen R, Aebersold R, Brentnall TA (January 2011). "Mass spectrometry based glycoproteomics--from a proteomics perspective". Molecular & Cellular Proteomics. 10 (1): R110.003251. doi: 10.1074/mcp.R110.003251 . PMC 3013464 . PMID 20736408.
- ↑ Singh A (January 2021). "Glycoproteomics". Nature Methods. 18 (1): 28. doi: 10.1038/s41592-020-01028-9 . PMID 33408388. S2CID 230796992.
- ↑ Liu H, Zhang N, Wan D, Cui M, Liu Z, Liu S (April 2014). "Mass spectrometry-based analysis of glycoproteins and its clinical applications in cancer biomarker discovery". Clinical Proteomics. 11 (1): 14. doi: 10.1186/1559-0275-11-14 . PMC 3984494 . PMID 24722010.