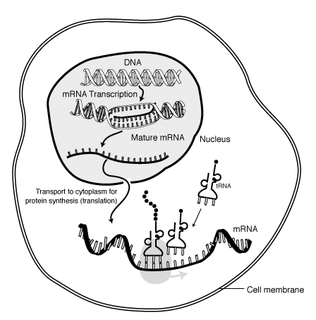
In molecular biology, messenger RNA (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.

Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product. These products are often proteins, but in non-protein-coding genes such as transfer RNA (tRNA) or small nuclear RNA (snRNA) genes, the product is a functional RNA. Gene expression is summarized in the central dogma of molecular biology first formulated by Francis Crick in 1958, further developed in his 1970 article, and expanded by the subsequent discoveries of reverse transcription and RNA replication.

In molecular genetics, the three prime untranslated region (3′-UTR) is the section of messenger RNA (mRNA) that immediately follows the translation termination codon. The 3′-UTR often contains regulatory regions that post-transcriptionally influence gene expression.
In molecular biology and genetics, transcriptional regulation is the means by which a cell regulates the conversion of DNA to RNA (transcription), thereby orchestrating gene activity. A single gene can be regulated in a range of ways, from altering the number of copies of RNA that are transcribed, to the temporal control of when the gene is transcribed. This control allows the cell or organism to respond to a variety of intra- and extracellular signals and thus mount a response. Some examples of this include producing the mRNA that encode enzymes to adapt to a change in a food source, producing the gene products involved in cell cycle specific activities, and producing the gene products responsible for cellular differentiation in multicellular eukaryotes, as studied in evolutionary developmental biology.
An oocyte, oöcyte, ovocyte, or rarely ocyte, is a female gametocyte or germ cell involved in reproduction. In other words, it is an immature ovum, or egg cell. An oocyte is produced in the ovary during female gametogenesis. The female germ cells produce a primordial germ cell (PGC), which then undergoes mitosis, forming oogonia. During oogenesis, the oogonia become primary oocytes. An oocyte is a form of genetic material that can be collected for cryoconservation. Cryoconservation of animal genetic resources has been put into action as a means of conserving traditional livestock.

Drosophila embryogenesis, the process by which Drosophila embryos form, is a favorite model system for genetics and developmental biology. The study of its embryogenesis unlocked the century-long puzzle of how development was controlled, creating the field of evolutionary developmental biology. The small size, short generation time, and large brood size make it ideal for genetic studies. Transparent embryos facilitate developmental studies. Drosophila melanogaster was introduced into the field of genetic experiments by Thomas Hunt Morgan in 1909.

A primary transcript is the single-stranded ribonucleic acid (RNA) product synthesized by transcription of DNA, and processed to yield various mature RNA products such as mRNAs, tRNAs, and rRNAs. The primary transcripts designated to be mRNAs are modified in preparation for translation. For example, a precursor mRNA (pre-mRNA) is a type of primary transcript that becomes a messenger RNA (mRNA) after processing.
Post-transcriptional modification or co-transcriptional modification is a set of biological processes common to most eukaryotic cells by which an RNA primary transcript is chemically altered following transcription from a gene to produce a mature, functional RNA molecule that can then leave the nucleus and perform any of a variety of different functions in the cell. There are many types of post-transcriptional modifications achieved through a diverse class of molecular mechanisms.
Ultrabithorax (Ubx) is a homeobox gene found in insects, and is used in the regulation of patterning in morphogenesis. There are many possible products of this gene, which function as transcription factors. Ubx is used in the specification of serially homologous structures, and is used at many levels of developmental hierarchies. In Drosophila melanogaster it is expressed in the third thoracic (T3) and first abdominal (A1) segments and represses wing formation. The Ubx gene regulates the decisions regarding the number of wings and legs the adult flies will have. The developmental role of the Ubx gene is determined by the splicing of its product, which takes place after translation of the gene. The specific splice factors of a particular cell allow the specific regulation of the developmental fate of that cell, by making different splice variants of transcription factors. In D. melanogaster, at least six different isoforms of Ubx exist.
In the field of developmental biology, regional differentiation is the process by which different areas are identified in the development of the early embryo. The process by which the cells become specified differs between organisms.

The bicoid 3′-UTR regulatory element is an mRNA regulatory element that controls the gene expression of the bicoid protein in fruitfly Drosophila melanogaster.
oskar is a gene required for the development of the Drosophila embryo. It defines the posterior pole during early embryogenesis. Its two isoforms, short and long, play different roles in Drosophila embryonic development.
The cytoplasmic polyadenylation element (CPE) is a sequence element found in the 3' untranslated region of messenger RNA. While several sequence elements are known to regulate cytoplasmic polyadenylation, CPE is the best characterized. The most common CPE sequence is UUUUAU, though there are other variations. Binding of CPE binding protein to this region promotes the extension of the existing polyadenine tail and, in general, activation of the mRNA for protein translation. This elongation occurs after the mRNA has been exported from the nucleus to the cytoplasm. A longer poly(A) tail attracts more cytoplasmic polyadenine binding proteins (PABPs) which interact with several other cytoplasmic proteins that encourage the mRNA and the ribosome to associate. The lengthening of the poly(A) tail thus has a role in increasing translational efficiency of the mRNA. The polyadenine tails are extended from approximately 40 bases to 150 bases.

Small Cajal body-specific RNAs (scaRNAs) are a class of small nucleolar RNAs (snoRNAs) that specifically localise to the Cajal body, a nuclear organelle involved in the biogenesis of small nuclear ribonucleoproteins. ScaRNAs guide the modification of RNA polymerase II transcribed spliceosomal RNAs U1, U2, U4, U5 and U12.
Vasa is an RNA binding protein with an ATP-dependent RNA helicase that is a member of the DEAD box family of proteins. The vasa gene, is essential for germ cell development and was first identified in Drosophila melanogaster, but has since been found to be conserved in a variety of vertebrates and invertebrates including humans. The Vasa protein is found primarily in germ cells in embryos and adults, where it is involved in germ cell determination and function, as well as in multipotent stem cells, where its exact function is unknown.

The developmentally active and heat shock inducible hsromega or hsrω gene in Drosophila produces multiple long non-coding RNA transcripts. This gene is transcriptionally active in almost all cell types of Drosophila and is the most actively induced following heat shock. A unique feature of the hsromega gene, which led to discovery of the 93D puff in 1970, is its singular inducibility with benzamide and a variety of other amides.
mir-279 is a short RNA molecule found in Drosophila melanogaster that belongs to a class of molecules known as microRNAs. microRNAs are ~22nt-long non-coding RNAs that post-transcriptionally regulate the expression of genes, often by binding to the 3' untranslated region of mRNA, targeting the transcript for degradation. miR-279 has diverse tissue-specific functions in the fly, influencing developmental processes related to neurogenesis and oogenesis, as well as behavioral processes related to circadian rhythms. The varied roles of mir-279, both in the developing and adult fly, highlight the utility of microRNAs in regulating unique biological processes.
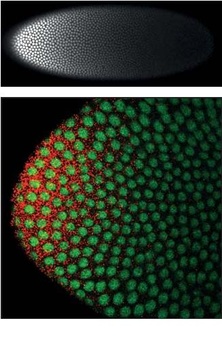
Bicoid is a maternal effect gene whose protein concentration gradient patterns the anterior-posterior (A-P) axis during Drosophila embryogenesis. Bicoid was the first protein demonstrated to act as a morphogen. Although Bicoid is important for the development of Drosophila and other higher dipterans, it is absent from most other insects, where its role is accomplished by other genes.

Smaug is a RNA binding protein in Drosophila that helps in maternal to zygotic transition (MZT). The protein is named after the fictional character Smaug, the dragon in J.R.R. Tolkien's 1937 novel The Hobbit. The MZT ends with the midblastula transition (MBT), which is defined as the first developmental event in Drosophila that depends on zygotic mRNA. In Drosophila, the initial developmental events are controlled by maternal mRNAs like Hsp83, nanos, string, Pgc, and cyclin B mRNA. Degradation of these mRNAs, which is expected to terminate maternal control and enable zygotic control of embryogenesis, happens at interphase of nuclear division cycle 14. During this transition smaug protein targets the maternal mRNA for destruction using miRs. Thus activating the zygotic genes. Smaug is expected to play a role in expression of three miRNAs – miR-3, miR-6, miR-309 and miR-286 during MZT in Drosophila. Among them smaug dependent expression of miR-309 is needed for destabilization of 410 maternal mRNA. In smaug mutants almost 85% of maternal mRNA is found to be stable. Smaug also bind to 3′ untranslated region (UTR) elements known as SMG response elements (SREs) on nanos mRNA and represses its expression by recruiting a protein called Cup(an eIF4E-binding protein that blocks the binding of eIF4G to eIF4E). There after it recruits deadenylation complex CCR4-Not on to the nanos mRNA which leads to deadenylation and subsequent decay of the mRNA. It is also found to be involved in degradation and repression of maternal Hsp83 mRNA by recruiting CCR4/POP2/NOT deadenylase to the mRNA. The human Smaug protein homologs are SAMD4A and SAMD4B.
Anne Ephrussi is a French developmental and molecular biologist. Her research is focussed on the study of post-transcriptional regulations such as mRNA localization and translation control in molecular biology as well as the establishment of polarity axes in cell and developmental biology. She is Head of the Developmental Biology Unit and director of the EMBL International Centre for Advanced Training (EICAT) program at the European Molecular Biology Laboratory (EMBL).