Enterobacteria phage λ is a bacterial virus, or bacteriophage, that infects the bacterial species Escherichia coli. It was discovered by Esther Lederberg in 1950 when she noticed that streaks of mixtures of two E. coli strains, one of which treated with ultraviolet light, was "nibbled and plaqued". The wild type of this virus has a temperate lifecycle that allows it to either reside within the genome of its host through lysogeny or enter into a lytic phase ; mutant strains are unable to lysogenize cells – instead, they grow and enter the lytic cycle after superinfecting an already lysogenized cell.

The lac operon is an operon required for the transport and metabolism of lactose in Escherichia coli and many other enteric bacteria. Although glucose is the preferred carbon source for most bacteria, the lac operon allows for the effective digestion of lactose when glucose is not available through the activity of beta-galactosidase. Gene regulation of the lac operon was the first genetic regulatory mechanism to be understood clearly, so it has become a foremost example of prokaryotic gene regulation. It is often discussed in introductory molecular and cellular biology classes for this reason. This lactose metabolism system was used by François Jacob and Jacques Monod to determine how a biological cell knows which enzyme to synthesize. Their work on the lac operon won them the Nobel Prize in Physiology in 1965.
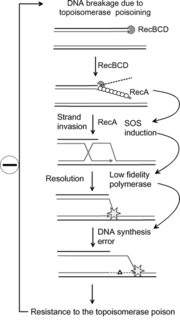
The SOS response is a global response to DNA damage in which the cell cycle is arrested and DNA repair and mutagenesis is induced. The system involves the RecA protein. The RecA protein, stimulated by single-stranded DNA, is involved in the inactivation of the repressor (LexA) of SOS response genes thereby inducing the response. It is an error-prone repair system that contributes significantly to DNA changes observed in a wide range of species.
Bacteriophage T7 is a bacteriophage, a virus that infects susceptible bacterial cells, that is composed of DNA and infects most strains of Escherichia coli. Bacteriophage T7 has a lytic life cycle and several properties that make it an ideal phage for experimentation.

3-Amino-1,2,4-triazole (3-AT) is a heterocyclic organic compound that consists of a 1,2,4-triazole substituted with an amino group.
HIS-selective medium is a type cell culture medium that lacks the amino acid histidine. It can be used with bacteria reliant on the expression of a gene encoding proteins involved in histidine expression in order to survive. Only bacteria expressing such genes will survive on these media.
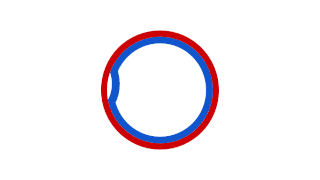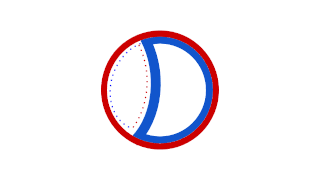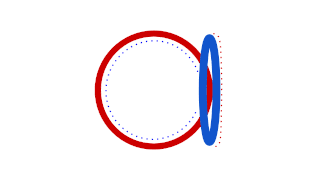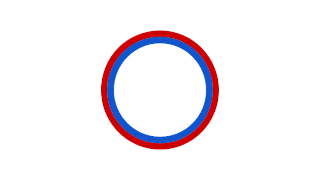
Prokaryotic DNA replication is the process by which a prokaryote duplicates its DNA into another copy that is passed on to daughter cells. Although it is often studied in the model organism E. coli, other bacteria show many similarities. Replication is bi-directional and originates at a single origin of replication (OriC). It consists of three steps: Initiation, elongation, and termination.
Maltose-binding protein (MBP) is a part of the maltose/maltodextrin system of Escherichia coli, which is responsible for the uptake and efficient catabolism of maltodextrins. It is a complex regulatory and transport system involving many proteins and protein complexes. MBP has an approximate molecular mass of 42.5 kilodaltons.
fis is an E. coli gene encoding the Fis protein. The regulation of this gene is more complex than most other genes in the E. coli genome, as Fis is an important protein which regulates expression of other genes. It is supposed that fis is regulated by H-NS, IHF and CRP. It also regulates its own expression (autoregulation). Fis is one of the most abundant DNA binding proteins in Escherichia coli under nutrient-rich growth conditions.
The degradosome is a multiprotein complex present in most bacteria that is involved in the processing of ribosomal RNA and the degradation of messenger RNA and is regulated by Non-coding RNA. It contains the proteins RNA helicase B, RNase E and Polynucleotide phosphorylase.
In molecular cloning, a vector is a DNA molecule used as a vehicle to artificially carry foreign genetic material into another cell, where it can be replicated and/or expressed. A vector containing foreign DNA is termed recombinant DNA. The four major types of vectors are plasmids, viral vectors, cosmids, and artificial chromosomes. Of these, the most commonly used vectors are plasmids. Common to all engineered vectors are an origin of replication, a multicloning site, and a selectable marker.

The bacterial one-hybrid (B1H) system is a method for identifying the sequence-specific target site of a DNA-binding domain. In this system, a given transcription factor (TF) is expressed as a fusion to a subunit of RNA polymerase. In parallel, a library of randomized oligonucleotides representing potential TF target sequences are cloned into a separate vector containing the selectable genes HIS3 and URA3. If the DNA-binding domain (bait) binds a potential DNA target site (prey) in vivo, it will recruit RNA polymerase to the promoter and activate transcription of the reporter genes in that clone. The two reporter genes, HIS3 and URA3, allow for positive and negative selections, respectively. At the end of the process, positive clones are sequenced and examined with motif-finding tools in order to resolve the favoured DNA target sequence.

A circular prokaryote chromosome is a chromosome in bacteria and archaea, in the form of a molecule of circular DNA. Unlike the linear DNA of most eukaryotes, typical prokaryote chromosomes are circular.

A toxin-antitoxin system is a set of two or more closely linked genes that together encode both a "toxin" protein and a corresponding "antitoxin". When these systems are contained on plasmids – transferable genetic elements – they ensure that only the daughter cells that inherit the plasmid survive after cell division. If the plasmid is absent in a daughter cell, the unstable antitoxin is degraded and the stable toxic protein kills the new cell; this is known as 'post-segregational killing' (PSK). Toxin-antitoxin systems are widely distributed in prokaryotes, and organisms often have them in multiple copies.

Escherichia coli is a Gram-negative gammaproteobacterium commonly found in the lower intestine of warm-blooded organisms (endotherms). The descendants of two isolates, K-12 and B strain, are used routinely in molecular biology as both a tool and a model organism.
In molecular biology, bacterial DNA binding proteins are a family of small, usually basic proteins of about 90 residues that bind DNA and are known as histone-like proteins. Since bacterial binding proteins have a diversity of functions, it has been difficult to develop a common function for all of them. They are commonly referred to as histone-like and have many similar traits with the eukaryotic histone proteins. Eukaryotic histones package DNA to help it to fit in the nucleus, and they are known to be the most conserved proteins in nature. Examples include the HU protein in Escherichia coli, a dimer of closely related alpha and beta chains and in other bacteria can be a dimer of identical chains. HU-type proteins have been found in a variety of eubacteria and archaebacteria, and are also encoded in the chloroplast genome of some algae. The integration host factor (IHF), a dimer of closely related chains which is suggested to function in genetic recombination as well as in translational and transcriptional control is found in Enterobacteria and viral proteins including the African swine fever virus protein A104R.
In DNA repair, the Ada Regulon is a set of genes whose expression is essential to adaptive response, which is triggered in prokaryotic cells by exposure to sub-lethal doses of alkylating agents. This allows the cells to tolerate the effects of such agents, which are otherwise toxic and mutagenic.

The Tac-Promoter, or tac vector is a synthetically produced DNA promoter, produced from the combination of promoters from the trp and lac operons. It is commonly used for protein production in Escherichia coli, Pichia pastoris and similar analytical operations including the ones leading to processes like molecular cloning.














