Related Research Articles

In the fields of molecular biology and genetics, a genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA. The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as regulatory sequences, and often a substantial fraction of 'junk' DNA with no evident function. Almost all eukaryotes have mitochondria and a small mitochondrial genome. Algae and plants also contain chloroplasts with a chloroplast genome.
A bacterial artificial chromosome (BAC) is a DNA construct, based on a functional fertility plasmid, used for transforming and cloning in bacteria, usually E. coli. F-plasmids play a crucial role because they contain partition genes that promote the even distribution of plasmids after bacterial cell division. The bacterial artificial chromosome's usual insert size is 150–350 kbp. A similar cloning vector called a PAC has also been produced from the DNA of P1 bacteriophage.

A generegulatory network (GRN) is a collection of molecular regulators that interact with each other and with other substances in the cell to govern the gene expression levels of mRNA and proteins which, in turn, determine the function of the cell. GRN also play a central role in morphogenesis, the creation of body structures, which in turn is central to evolutionary developmental biology (evo-devo).

FtsZ is a protein encoded by the ftsZ gene that assembles into a ring at the future site of bacterial cell division. FtsZ is a prokaryotic homologue of the eukaryotic protein tubulin. The initials FtsZ mean "Filamenting temperature-sensitive mutant Z." The hypothesis was that cell division mutants of E. coli would grow as filaments due to the inability of the daughter cells to separate from one another. FtsZ is found in almost all bacteria, many archaea, all chloroplasts and some mitochondria, where it is essential for cell division. FtsZ assembles the cytoskeletal scaffold of the Z ring that, along with additional proteins, constricts to divide the cell in two.

Caulobacter crescentus is a Gram-negative, oligotrophic bacterium widely distributed in fresh water lakes and streams. The taxon is more properly known as Caulobacter vibrioides.

Synthetic biology (SynBio) is a multidisciplinary field of science that focuses on living systems and organisms, and it applies engineering principles to develop new biological parts, devices, and systems or to redesign existing systems found in nature.
Radiosensitivity is the relative susceptibility of cells, tissues, organs or organisms to the harmful effect of ionizing radiation.

Angiogenin (ANG) also known as ribonuclease 5 is a small 123 amino acid protein that in humans is encoded by the ANG gene. Angiogenin is a potent stimulator of new blood vessels through the process of angiogenesis. Ang hydrolyzes cellular RNA, resulting in modulated levels of protein synthesis and interacts with DNA causing a promoter-like increase in the expression of rRNA. Ang is associated with cancer and neurological disease through angiogenesis and through activating gene expression that suppresses apoptosis.

Gail Roberta Martin is an American biologist. She is professor emerita in the Department of Anatomy, University of California, San Francisco. She is known for her pioneering work on the isolation of pluripotent stem cells from normal embryos, for which she coined the term ‘embryonic stem cells’. She is also widely recognized for her work on the function of Fibroblast Growth Factors (FGFs) and their negative regulators in vertebrate organogenesis. She and her colleagues also made valuable contributions to gene targeting technology.

A prokaryote is a single-celled organism that lacks a nucleus and other membrane-bound organelles. The word prokaryote comes from the Greek πρό and κάρυον. In the two-empire system arising from the work of Édouard Chatton, prokaryotes were classified within the empire Prokaryota. But in the three-domain system, based upon molecular analysis, prokaryotes are divided into two domains: Bacteria and Archaea. Organisms with nuclei are placed in a third domain, Eukaryota. In biological evolution, prokaryotes are deemed to have arisen before eukaryotes.
Transcriptional noise is a primary cause of the variability (noise) in gene expression occurring between cells in isogenic populations. A proposed source of transcriptional noise is transcriptional bursting although other sources of heterogeneity, such as unequal separation of cell contents at mitosis are also likely to contribute considerably. Bursting transcription, as opposed to simple probabilistic models of transcription, reflects multiple states of gene activity, with fluctuations between states separated by irregular intervals, generating uneven protein expression between cells. Noise in gene expression can have tremendous consequences on cell behaviour, and must be mitigated or integrated. In certain contexts, such as establishment of viral latency, the survival of microbes in rapidly changing stressful environments, or several types of scattered differentiation, the variability may be essential. Variability also impacts upon the effectiveness of clinical treatment, with resistance of bacteria and yeast to antibiotics demonstrably caused by non-genetic differences. Variability in gene expression may also contribute to resistance of sub-populations of cancer cells to chemotherapy and appears to be a barrier to curing HIV.
Michael B. Elowitz is a biologist and professor of Biology, Bioengineering, and Applied Physics at the California Institute of Technology, and investigator at the Howard Hughes Medical Institute. In 2007 he was the recipient of the Genius grant, better known as the MacArthur Fellows Program for the design of a synthetic gene regulatory network, the Repressilator, which helped initiate the field of synthetic biology. In addition, he showed, for the first time, how inherently random effects, or 'noise', in gene expression could be detected and quantified in living cells, leading to a growing recognition of the many roles that noise plays in living cells. His work in Synthetic Biology and Noise represent two foundations of the field of Systems Biology.
Xiaowei Zhuang is a Chinese-American biophysicist who is the David B. Arnold Jr. Professor of Science, Professor of Chemistry and Chemical Biology, and Professor of Physics at Harvard University, and an Investigator at the Howard Hughes Medical Institute. She is best known for her work in the development of Stochastic Optical Reconstruction Microscopy (STORM), a super-resolution fluorescence microscopy method, and the discoveries of novel cellular structures using STORM. She received a 2019 Breakthrough Prize in Life Sciences for developing super-resolution imaging techniques that get past the diffraction limits of traditional light microscopes, allowing scientists to visualize small structures within living cells. She was elected a Member of the American Philosophical Society in 2019 and was awarded a Vilcek Foundation Prize in Biomedical Science in 2020.

In evolutionary biology, robustness of a biological system is the persistence of a certain characteristic or trait in a system under perturbations or conditions of uncertainty. Robustness in development is known as canalization. According to the kind of perturbation involved, robustness can be classified as mutational, environmental, recombinational, or behavioral robustness etc. Robustness is achieved through the combination of many genetic and molecular mechanisms and can evolve by either direct or indirect selection. Several model systems have been developed to experimentally study robustness and its evolutionary consequences.
Cellular noise is random variability in quantities arising in cellular biology. For example, cells which are genetically identical, even within the same tissue, are often observed to have different expression levels of proteins, different sizes and structures. These apparently random differences can have important biological and medical consequences.
Johan Paulsson is a Swedish mathematician and systems biologist at Harvard Medical School. He is a leading researcher in systems biology and stochastic processes, specializing in stochasticity in gene networks and plasmid reproduction.

Synthetic biological circuits are an application of synthetic biology where biological parts inside a cell are designed to perform logical functions mimicking those observed in electronic circuits. The applications range from simply inducing production to adding a measurable element, like GFP, to an existing natural biological circuit, to implementing completely new systems of many parts.

Lucy Shapiro is an American developmental biologist. She is a professor of Developmental Biology at the Stanford University School of Medicine. She is the Virginia and D.K. Ludwig Professor of Cancer Research and the director of the Beckman Center for Molecular and Genetic Medicine.
The parABS system is a broadly conserved molecular mechanism for plasmid partitioning and chromosome segregation in bacteria. Originally identified as a genetic element required for faithful partitioning of low-copy-number plasmids, it consists of three components: the ParA ATPase, the ParB DNA-binding protein, and the cis-acting parS sequence. The parA and parB genes are typically found in the same operon, with parS elements located within or adjacent to this operon. Collectively, these components function to ensure accurate partitioning of plasmids or whole chromosomes between bacterial daughter cells prior to cell division.
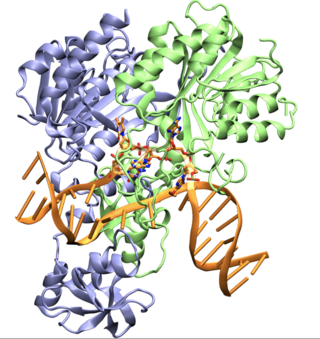
CcrM is an orphan DNA methyltransferase, that is involved in controlling gene expression in most Alphaproteobacteria. This enzyme modifies DNA by catalyzing the transference of a methyl group from the S-adenosyl-L methionine substrate to the N6 position of an adenine base in the sequence 5'-GANTC-3' with high specificity. In some lineages such as SAR11, the homologous enzymes possess 5'-GAWTC-3' specificity. In Caulobacter crescentus Ccrm is produced at the end of the replication cycle when Ccrm recognition sites are hemimethylated, rapidly methylating the DNA. CcrM is essential in other Alphaproteobacteria but is role is not yet determined. CcrM is a highly specific methyltransferase with a novel DNA recognition mechanism.
References
- ↑ McAdams, H. H.; Shapiro, L. (1995). "Circuit simulation of genetic networks". Science . 269 (5224): 650–6. Bibcode:1995Sci...269..650M. doi:10.1126/science.7624793. PMID 7624793.
- ↑ Arkin, A.; Ross, J.; McAdams, H. H. (1998). "Stochastic kinetic analysis of developmental pathway bifurcation in phage lambda-infected Escherichia coli cells". Genetics . 149 (4): 1633–48. doi:10.1093/genetics/149.4.1633. PMC 1460268 . PMID 9691025.
- ↑ McAdams, H. H.; Arkin, A. (1998). "Simulation of prokaryotic genetic circuits". Annual Review of Biophysics and Biomolecular Structure. 27: 199–224. doi:10.1146/annurev.biophys.27.1.199. PMID 9646867.
- ↑ McAdams, H. H.; Arkin, A. (1999). "It's a noisy business! Genetic regulation at the nanomolar scale". Trends Genet. 15 (2): 65–69. doi:10.1016/S0168-9525(98)01659-X. PMID 10098409.
- ↑ McAdams, H. H.; Arkin, A. (1997). "Stochastic mechanisms in gene expression". Proc. Natl. Acad. Sci. U.S.A. 94 (3): 814–9. Bibcode:1997PNAS...94..814M. doi: 10.1073/pnas.94.3.814 . PMC 19596 . PMID 9023339.
- ↑ McAdams, H. H.; Shapiro, L. (2009). "System-level design of bacterial cell cycle control". FEBS Lett. 586 (2): 3984–3991. doi:10.1016/j.febslet.2009.09.030. PMC 2795017 . PMID 19766635.
- ↑ McAdams, H. H.; Shapiro, L. (2003). "A bacterial cell-cycle regulatory network operating in time and space". Science . 301 (5641): 3984–3991. Bibcode:2003Sci...301.1874M. doi:10.1126/science.1087694. PMID 14512618. S2CID 28742912.
- ↑ "The John Scott Award".
- ↑ "Stanford Profile, Harley H McAdams".