Related Research Articles

Ribonucleic acid (RNA) is a polymeric molecule that is essential for most biological functions, either by performing the function itself or by forming a template for production of proteins. RNA and deoxyribonucleic acid (DNA) are nucleic acids. The nucleic acids constitute one of the four major macromolecules essential for all known forms of life. RNA is assembled as a chain of nucleotides. Cellular organisms use messenger RNA (mRNA) to convey genetic information that directs synthesis of specific proteins. Many viruses encode their genetic information using an RNA genome.
Hepatitis D is a type of viral hepatitis caused by the hepatitis delta virus (HDV). HDV is one of five known hepatitis viruses: A, B, C, D, and E. HDV is considered to be a satellite because it can propagate only in the presence of the hepatitis B virus (HBV). Transmission of HDV can occur either via simultaneous infection with HBV (coinfection) or superimposed on chronic hepatitis B or hepatitis B carrier state (superinfection).

Transfer RNA is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length, that serves as the physical link between the mRNA and the amino acid sequence of proteins. Transfer RNA (tRNA) does this by carrying an amino acid to the protein synthesizing machinery of a cell called the ribosome. Complementation of a 3-nucleotide codon in a messenger RNA (mRNA) by a 3-nucleotide anticodon of the tRNA results in protein synthesis based on the mRNA code. As such, tRNAs are a necessary component of translation, the biological synthesis of new proteins in accordance with the genetic code.

Transcriptional modification or co-transcriptional modification is a set of biological processes common to most eukaryotic cells by which an RNA primary transcript is chemically altered following transcription from a gene to produce a mature, functional RNA molecule that can then leave the nucleus and perform any of a variety of different functions in the cell. There are many types of post-transcriptional modifications achieved through a diverse class of molecular mechanisms.

A long terminal repeat (LTR) is a pair of identical sequences of DNA, several hundred base pairs long, which occur in eukaryotic genomes on either end of a series of genes or pseudogenes that form a retrotransposon or an endogenous retrovirus or a retroviral provirus. All retroviral genomes are flanked by LTRs, while there are some retrotransposons without LTRs. Typically, an element flanked by a pair of LTRs will encode a reverse transcriptase and an integrase, allowing the element to be copied and inserted at a different location of the genome. Copies of such an LTR-flanked element can often be found hundreds or thousands of times in a genome. LTR retrotransposons comprise about 8% of the human genome.

The hepatitis delta virus (HDV) ribozyme is a non-coding RNA found in the hepatitis delta virus that is necessary for viral replication and is the only known human virus that utilizes ribozyme activity to infect its host. The ribozyme acts to process the RNA transcripts to unit lengths in a self-cleavage reaction during replication of the hepatitis delta virus, which is thought to propagate by a double rolling circle mechanism. The ribozyme is active in vivo in the absence of any protein factors and was the fastest known naturally occurring self-cleaving RNA at the time of its discovery.

The retroviral psi packaging element, also known as the Ψ RNA packaging signal, is a cis-acting RNA element identified in the genomes of the retroviruses Human immunodeficiency virus (HIV) and Simian immunodeficiency virus (SIV). It is involved in regulating the essential process of packaging the retroviral RNA genome into the viral capsid during replication. The final virion contains a dimer of two identical unspliced copies of the viral genome.

U1 spliceosomal RNA is the small nuclear RNA (snRNA) component of U1 snRNP, an RNA-protein complex that combines with other snRNPs, unmodified pre-mRNA, and various other proteins to assemble a spliceosome, a large RNA-protein molecular complex upon which splicing of pre-mRNA occurs. Splicing, or the removal of introns, is a major aspect of post-transcriptional modification, and takes place only in the nucleus of eukaryotes.

Heterogeneous nuclear ribonucleoprotein A1 is a protein that in humans is encoded by the HNRNPA1 gene. Mutations in hnRNP A1 are causative of amyotrophic lateral sclerosis and the syndrome multisystem proteinopathy.

Poly(rC)-binding protein 2 is a protein that in humans is encoded by the PCBP2 gene.

SON protein is a protein that in humans is encoded by the SON gene.

MHC class II regulatory factor RFX1 is a protein that, in humans, is encoded by the RFX1 gene located on the short arm of chromosome 19.
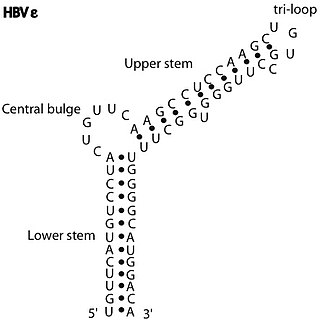
The HBV RNA encapsidation signal epsilon is an element essential for HBV virus replication.
The Hepatitis B virus PRE stem-loop beta is an RNA structure that is shown to play a role in nuclear export of HBV mRNAs.
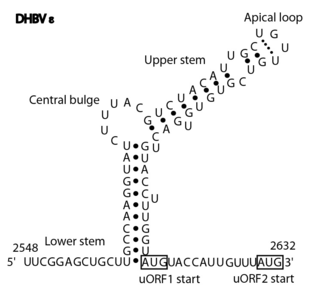
The Avian HBV RNA encapsidation signal epsilon is an RNA structure that is shown to facilitate encapsidation of the pregenomic RNA required for viral replication. There are two main classes of encapsidation signals in avian hepatitis B viruses - Duck hepatitis B virus (DHBV) and Heron hepatitis B virus (HHBV) like. DHBV is used as a model to understand human Hepatitis B virus. Although studies have shown that the HHBV epsilon has less pairing in the upper stem than DHBV, this pairing is not absolutely required for DHBV infection in ducks.

Rev is a transactivating protein that is essential to the regulation of HIV-1 protein expression. A nuclear localization signal is encoded in the rev gene, which allows the Rev protein to be localized to the nucleus, where it is involved in the export of unspliced and incompletely spliced mRNAs. In the absence of Rev, mRNAs of the HIV-1 late (structural) genes are retained in the nucleus, preventing their translation.

Hepatitis B virus (HBV) is a partially double-stranded DNA virus, a species of the genus Orthohepadnavirus and a member of the Hepadnaviridae family of viruses. This virus causes the disease hepatitis B.

Nucleic acid secondary structure is the basepairing interactions within a single nucleic acid polymer or between two polymers. It can be represented as a list of bases which are paired in a nucleic acid molecule. The secondary structures of biological DNAs and RNAs tend to be different: biological DNA mostly exists as fully base paired double helices, while biological RNA is single stranded and often forms complex and intricate base-pairing interactions due to its increased ability to form hydrogen bonds stemming from the extra hydroxyl group in the ribose sugar.

HSURs are viral small regulatory RNAs. They are found in Herpesvirus saimiri which is responsible for aggressive T-cell leukemias in primates. They are nuclear RNAs which bind host proteins to form small nuclear ribonucleoproteins (snRNPs). The RNAs are 114–143 nucleotides in length and the HSUR family has been subdivided into HSURs numbered 1 to 7. The function of HSURs has not yet been identified; they do not affect transcription so are thought to act post-transcriptionally, potentially influencing the stability of host mRNAs.
Hepatitis B virus PRE 1151–1410 is a part of 500 base pair long HBV PRE, that has been proposed to be the hepatitis B virus (HBV) RNA export element. However, the function is controversial and new regulatory elements have been predicted within PRE. PRE 1151–1410 enhances nuclear export of intronless transcripts and represses the splicing mechanism to a comparable degree to that of the full-length PRE. Hence it was proposed to be the core HBV PRE element. PRE1151–1410 contains 3 known regulatory elements: PRE SL-alpha, human La protein binding site, SRE-1.
References
- ↑ Smith Gj, 3rd; Donello, JE; Lück, R; Steger, G; Hope, TJ (1998). "The hepatitis B virus post-transcriptional regulatory element contains two conserved RNA stem-loops which are required for function". Nucleic Acids Research. 26 (21): 4818–4827. doi:10.1093/nar/26.21.4818. PMC 147918 . PMID 9776740.
- ↑ Schwalbe, M; Ohlenschläger, O; Marchanka, A; Ramachandran, R; Häfner, S; Heise, T; Görlach, M (March 2008). "Solution structure of stem-loop alpha of the hepatitis B virus post-transcriptional regulatory element". Nucleic Acids Research. 36 (5): 1681–1689. doi:10.1093/nar/gkn006. PMC 2275152 . PMID 18263618.
- ↑ Lim, CS; Brown, CM (September 2016). "Hepatitis B virus nuclear export elements: RNA stem-loop α and β, key parts of the HBV post-transcriptional regulatory element". RNA Biology. 13 (9): 743–747. doi:10.1080/15476286.2016.1166330. PMC 5013995 . PMID 27031749.
- ↑ Chen, A; Panjaworayan T-Thienprasert, N; Brown, CM (7 July 2014). "Prospects for inhibiting the post-transcriptional regulation of gene expression in hepatitis B virus". World Journal of Gastroenterology. 20 (25): 7993–8004. doi: 10.3748/wjg.v20.i25.7993 . PMC 4081668 . PMID 25009369.