Related Research Articles
Hepatitis D is a type of viral hepatitis caused by the hepatitis delta virus (HDV). HDV is one of five known hepatitis viruses: A, B, C, D, and E. HDV is considered to be a satellite because it can propagate only in the presence of the hepatitis B virus (HBV). Transmission of HDV can occur either via simultaneous infection with HBV (coinfection) or superimposed on chronic hepatitis B or hepatitis B carrier state (superinfection).

Hepadnaviridae is a family of viruses. Humans, apes, and birds serve as natural hosts. There are currently 18 species in this family, divided among 5 genera. Its best-known member is hepatitis B virus. Diseases associated with this family include: liver infections, such as hepatitis, hepatocellular carcinomas, and cirrhosis. It is the sole accepted family in the order Blubervirales.

A long terminal repeat (LTR) is a pair of identical sequences of DNA, several hundred base pairs long, which occur in eukaryotic genomes on either end of a series of genes or pseudogenes that form a retrotransposon or an endogenous retrovirus or a retroviral provirus. All retroviral genomes are flanked by LTRs, while there are some retrotransposons without LTRs. Typically, an element flanked by a pair of LTRs will encode a reverse transcriptase and an integrase, allowing the element to be copied and inserted at a different location of the genome. Copies of such an LTR-flanked element can often be found hundreds or thousands of times in a genome. LTR retrotransposons comprise about 8% of the human genome.

The hepatitis delta virus (HDV) ribozyme is a non-coding RNA found in the hepatitis delta virus that is necessary for viral replication and is the only known human virus that utilizes ribozyme activity to infect its host. The ribozyme acts to process the RNA transcripts to unit lengths in a self-cleavage reaction during replication of the hepatitis delta virus, which is thought to propagate by a double rolling circle mechanism. The ribozyme is active in vivo in the absence of any protein factors and was the fastest known naturally occurring self-cleaving RNA at the time of its discovery.

The retroviral psi packaging element, also known as the Ψ RNA packaging signal, is a cis-acting RNA element identified in the genomes of the retroviruses Human immunodeficiency virus (HIV) and Simian immunodeficiency virus (SIV). It is involved in regulating the essential process of packaging the retroviral RNA genome into the viral capsid during replication. The final virion contains a dimer of two identical unspliced copies of the viral genome.

U1 spliceosomal RNA is the small nuclear RNA (snRNA) component of U1 snRNP, an RNA-protein complex that combines with other snRNPs, unmodified pre-mRNA, and various other proteins to assemble a spliceosome, a large RNA-protein molecular complex upon which splicing of pre-mRNA occurs. Splicing, or the removal of introns, is a major aspect of post-transcriptional modification, and takes place only in the nucleus of eukaryotes.

Hepatitis C alternative reading frame stem-loop is a conserved secondary structure motif identified in the RNA genome of the Hepatitis C virus (HCV) which is proposed to have an important role in regulating translation and repression of the viral genome.

SON protein is a protein that in humans is encoded by the SON gene.

Hepatitis B is an infectious disease caused by the Hepatitis B virus (HBV) that affects the liver; it is a type of viral hepatitis. It can cause both acute and chronic infection.
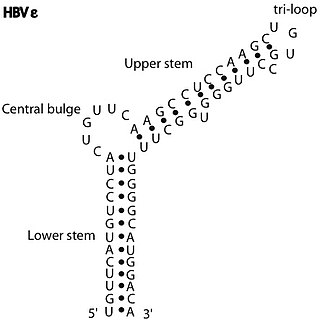
The HBV RNA encapsidation signal epsilon is an element essential for HBV virus replication.
The Hepatitis B virus PRE stem-loop alpha is an RNA structure that is shown to play a role in nuclear export of HBV mRNAs.
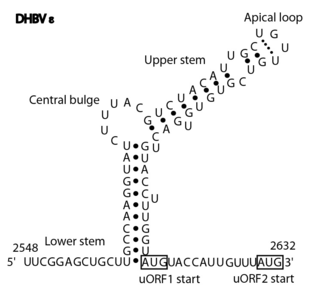
The Avian HBV RNA encapsidation signal epsilon is an RNA structure that is shown to facilitate encapsidation of the pregenomic RNA required for viral replication. There are two main classes of encapsidation signals in avian hepatitis B viruses - Duck hepatitis B virus (DHBV) and Heron hepatitis B virus (HHBV) like. DHBV is used as a model to understand human Hepatitis B virus. Although studies have shown that the HHBV epsilon has less pairing in the upper stem than DHBV, this pairing is not absolutely required for DHBV infection in ducks.

HBx is a hepatitis B viral protein. It is 154 amino acids long and interferes with transcription, signal transduction, cell cycle progress, protein degradation, apoptosis and chromosomal stability in the host. It forms a heterodimeric complex with its cellular target protein, and this interaction dysregulates centrosome dynamics and mitotic spindle formation. It interacts with DDB1 redirecting the ubiquitin ligase activity of the CUL4-DDB1 E3 complexes, which are intimately involved in the intracellular regulation of DNA replication and repair, transcription and signal transduction.

Hepatitis B virus DNA polymerase is a hepatitis B viral protein. It is a DNA polymerase that can use either DNA or RNA templates and a ribonuclease H that cuts RNA in the duplex. Both functions are supplied by the reverse transcriptase (RT) domain.

Hepatitis B virus (HBV) is a partially double-stranded DNA virus, a species of the genus Orthohepadnavirus and a member of the Hepadnaviridae family of viruses. This virus causes the disease hepatitis B.
cccDNA is a special DNA structure that arises during the propagation of some viruses in the cell nucleus and may remain permanently there. It is a double-stranded DNA that originates in a linear form that is ligated by means of DNA ligase to a covalently closed ring. In most cases, transcription of viral DNA can occur from the circular form only. The cccDNA of viruses is also known as episomal DNA or occasionally as a minichromosome.
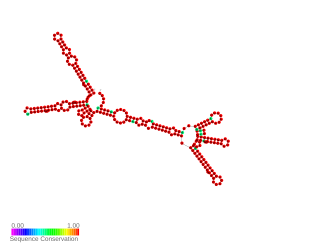
Hepatitis B virus PRE 1151–1410 is a part of 500 base pair long HBV PRE, that has been proposed to be the hepatitis B virus (HBV) RNA export element. However, the function is controversial and new regulatory elements have been predicted within PRE. PRE 1151–1410 enhances nuclear export of intronless transcripts and represses the splicing mechanism to a comparable degree to that of the full-length PRE. Hence it was proposed to be the core HBV PRE element. PRE1151–1410 contains 3 known regulatory elements: PRE SL-alpha, human La protein binding site, SRE-1.

Ground squirrel hepatitis virus, abbreviated GSHV, is a partially double-stranded DNA virus that is closely related to human Hepatitis B virus (HBV) and Woodchuck hepatitis virus (WHV). It is a member of the family of viruses Hepadnaviridae and the genus Orthohepadnavirus. Like the other members of its family, GSHV has high degree of species and tissue specificity. It was discovered in Beechey ground squirrels, Spermophilus beecheyi, but also infects Arctic ground squirrels, Spermophilus parryi. Commonalities between GSHV and HBV include morphology, DNA polymerase activity in genome repair, cross-reacting viral antigens, and the resulting persistent infection with viral antigen in the blood (antigenemia). As a result, GSHV is used as an experimental model for HBV.

The woolly monkey hepatitis B virus (WMHBV) is a viral species of the Orthohepadnavirus genus of the Hepadnaviridae family. Its natural host is the woolly monkey (Lagothrix), an inhabitant of South America categorized as a New World primate. WMHBV, like other hepatitis viruses, infects the hepatocytes, or liver cells, of its host organism. It can cause hepatitis, liver necrosis, cirrhosis, and hepatocellular carcinoma. Because nearly all species of Lagothrix are threatened or endangered, researching and developing a vaccine and/or treatment for WMHBV is important for the protection of the whole woolly monkey genus.
Coronavirus genomes are positive-sense single-stranded RNA molecules with an untranslated region (UTR) at the 5′ end which is called the 5′ UTR. The 5′ UTR is responsible for important biological functions, such as viral replication, transcription and packaging. The 5′ UTR has a conserved RNA secondary structure but different Coronavirus genera have different structural features described below.
References
- ↑ Smith Gj, 3rd; Donello, JE; Lück, R; Steger, G; Hope, TJ (1998). "The hepatitis B virus post-transcriptional regulatory element contains two conserved RNA stem-loops which are required for function". Nucleic Acids Research. 26 (21): 4818–4827. doi:10.1093/nar/26.21.4818. PMC 147918 . PMID 9776740.
- ↑ Chen, A; Panjaworayan T-Thienprasert, N; Brown, CM (7 July 2014). "Prospects for inhibiting the post-transcriptional regulation of gene expression in hepatitis B virus". World Journal of Gastroenterology. 20 (25): 7993–8004. doi: 10.3748/wjg.v20.i25.7993 . PMC 4081668 . PMID 25009369.