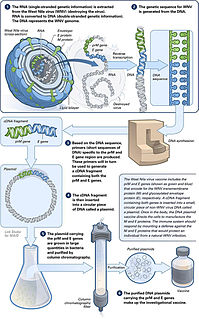
A DNA vaccine is a type of vaccine that transfects a specific antigen-coding DNA sequence into the cells of an organism as a mechanism to induce an immune response.

Luciferase is a generic term for the class of oxidative enzymes that produce bioluminescence, and is usually distinguished from a photoprotein. The name was first used by Raphaël Dubois who invented the words luciferin and luciferase, for the substrate and enzyme, respectively. Both words are derived from the Latin word lucifer, meaning "lightbearer", which in turn is derived from the Latin words for "light" (lux) and "to bring or carry" (ferre).
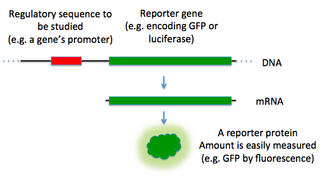
In molecular biology, a reporter gene is a gene that researchers attach to a regulatory sequence of another gene of interest in bacteria, cell culture, animals or plants. Such genes are called reporters because the characteristics they confer on organisms expressing them are easily identified and measured, or because they are selectable markers. Reporter genes are often used as an indication of whether a certain gene has been taken up by or expressed in the cell or organism population.

A prophage is a bacteriophage genome that is integrated into the circular bacterial chromosome or exists as an extrachromosomal plasmid within the bacterial cell. Integration of prophages into the bacterial host is the characteristic step of the lysogenic cycle of temperate phages. Prophages remain latent in the genome through multiple cell divisions until activation by an external factor, such as UV light, leading to production of new phage particles that will lyse the cell and spread. As ubiquitous mobile genetic elements, prophages play important roles in bacterial genetics and evolution, such as in the acquisition of virulence factors.

In molecular biology and genetics, transformation is the genetic alteration of a cell resulting from the direct uptake and incorporation of exogenous genetic material from its surroundings through the cell membrane(s). For transformation to take place, the recipient bacterium must be in a state of competence, which might occur in nature as a time-limited response to environmental conditions such as starvation and cell density, and may also be induced in a laboratory.

Escherichia virus T4 is a species of bacteriophages that infect Escherichia coli bacteria. It is a double-stranded DNA virus in the subfamily Tevenvirinae from the family Myoviridae. T4 is capable of undergoing only a lytic lifecycle and not the lysogenic lifecycle. The species was formerly named T-even bacteriophage, a name which also encompasses, among other strains, Enterobacteria phage T2, Enterobacteria phage T4 and Enterobacteria phage T6.
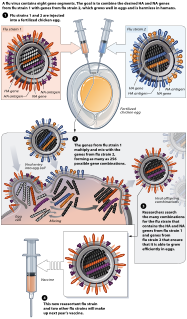
Reassortment is the mixing of the genetic material of a species into new combinations in different individuals. Several different processes contribute to reassortment, including assortment of chromosomes, and chromosomal crossover. It is particularly used when two similar viruses that are infecting the same cell exchange genetic material. In particular, reassortment occurs among influenza viruses, whose genomes consist of eight distinct segments of RNA. These segments act like mini-chromosomes, and each time a flu virus is assembled, it requires one copy of each segment.

Nucleotide excision repair is a DNA repair mechanism. DNA damage occurs constantly because of chemicals, radiation and other mutagens. Three excision repair pathways exist to repair single stranded DNA damage: Nucleotide excision repair (NER), base excision repair (BER), and DNA mismatch repair (MMR). While the BER pathway can recognize specific non-bulky lesions in DNA, it can correct only damaged bases that are removed by specific glycosylases. Similarly, the MMR pathway only targets mismatched Watson-Crick base pairs.

Two-hybrid screening is a molecular biology technique used to discover protein–protein interactions (PPIs) and protein–DNA interactions by testing for physical interactions between two proteins or a single protein and a DNA molecule, respectively.

Halobacterium is a genus in the family Halobacteriaceae.

Zearalenone (ZEN), also known as RAL and F-2 mycotoxin, is a potent estrogenic metabolite produced by some Fusarium and Gibberella species. Specifically, the Gibberella zeae, the fungal species where zearalenone was initially detected, in its asexual/anamorph stage is known as Fusarium graminearum. Several Fusarium species produce toxic substances of considerable concern to livestock and poultry producers, namely deoxynivalenol, T-2 toxin, HT-2 toxin, diacetoxyscirpenol (DAS) and zearalenone. Particularly, ZEN is produced by Fusarium graminearum, Fusarium culmorum, Fusarium cerealis, Fusarium equiseti, Fusarium verticillioides, and Fusarium incarnatum. Zearalenone is the primary toxin that binds to estrogen receptors, causing infertility, abortion or other breeding problems, especially in swine. Often, ZEN is detected together with deoxynivalenol in contaminated samples and its toxicity needs to be considered in combination with the presence of other toxins.

The single cell gel electrophoresis assay is an uncomplicated and sensitive technique for the detection of DNA damage at the level of the individual eukaryotic cell. It was first developed by Östling & Johansson in 1984 and later modified by Singh et al. in 1988. It has since increased in popularity as a standard technique for evaluation of DNA damage/repair, biomonitoring and genotoxicity testing. It involves the encapsulation of cells in a low-melting-point agarose suspension, lysis of the cells in neutral or alkaline (pH>13) conditions, and electrophoresis of the suspended lysed cells. The term "comet" refers to the pattern of DNA migration through the electrophoresis gel, which often resembles a comet.
A helper dependent virus, also termed a gutless virus, is a synthetic viral vector dependent on the assistance of a helper virus in order to replicate, and can be used for purposes such as gene therapy. Naturally-occurring satellite viruses are also helper virus dependent, and can sometimes be modified to become viral vectors.
UV-induced apoptosis UV-induced apoptosis is an adequate (physiological) reaction of a cell damaged by UV radiation (UVR) in a sufficiently large (lethal) dose and it prevents the disordered destruction of UV damaged cells by help necrosis. Cell elimination by apoptosis occurs when UV-induced cell damage which cannot be repaired by the intracellular repair system exceeds at it certain limit. Through apoptosis, the cells are self-disassembled into compartments with their subsequent utilization. The first time sign of the beginning of the apoptosis system is working in a UV damaged cell is the activation of restriction enzymes, which divide cell DNA into fragments convenient for utilization. But too large a dose of UVR can lead to breakdown (inactivation) of the energy-dependent mechanism of apoptosis. In this case, cell destruction occurs randomly, not orderly, and during a significantly longer time interval. UV-irradiated cells do not change their appearance for a long time [1, 6], as a result of which the researchers may make the erroneous conclusion that “revealed an unexpected response to a dose at which a higher dose of UV increased the viability of keratinocytes” [2]. The fact that UV-induced apoptosis at high doses of UVR begins to be replaced by necrosis was established in 2000 [3]. For keratinocytes, the proportion of cells that have elimination by help apoptosis, with an increase in UVR dose can reach to achieve 45%, but with a further increase in the dose of UVR, destruction of damaged cells by help necrosis and the part of cells that eliminated by apoptosis begins to decrease [4, 11]. In the dose range of UVR from “lethal” to “super-lethal”, “pro-inflammatory” apoptosis can be manifested, which was experimentally discovered in 2003 [5]. This may be the result of partial damage to the apoptosis mechanism by UV radiation [1]. If at moderate doses “pure” apoptosis does not cause an inflammatory reaction, then at sufficiently large doses, an inflammatory reaction arises due to pro-inflammatory apoptosis, which leads to the appearance of “fast” erythema for UV irradiated skin keratinocytes. Kinetic of “fast” erythema is much faster by the time of development of UV erythema caused by necrosis of UV damaged keratinocytes [6]. The most erythemogenic is UVB the spectral range of UVR, since radiation in this range is less absorbed by the outer layers of the skin, which allows UVB radiation, in contrast to UVC, to reach more deep layers skin and act on keratinocytes of the deep-lying basal layer of the epidermis of the skin. The ability to induce apoptosis for UVB and UVC radiation is due to the fact that the DNA of the nucleus [7] and / or mitochondria [8] of the cell absorbs UVR well in the UVC and UVB spectral range. Keratinocytes of the skin are in a state of programmed apoptosis, during which the keratinocytes of the basal layer are removed from it and during the transition through all layers of the epidermis within 28 days turn into flakes of the outer stratum corneum, which are subsequently desquamated. It is clear that the keratinocyte response to UV exposure will depend on what phase of programmed apoptosis the keratinocyte experienced UV exposure, and this is the main reason for the difference of the UV effect for UVC and UVB on the skin. There are also differences in the initiation of mitochondrial (internal) and caspase-dependent (external) apoptosis for the UVC and UVB spectral ranges [9]. Sunburn cells (SBS) are the keratinocytes in the process of UV-induced apoptosis. The appearance of SBC may be not associated with an inflammatory reaction, but the role of UV-induced apoptosis of skin keratinocytes in the development of UV erythema of the skin has been established, which allowed the development of a patent-protected METHOD FOR QUANTITATIVE ASSESSMENT OF APOPTOSIS SYSTEM [10], in which “the brightest lamp of skin display "(photoerythema) is used to diagnose the state of the body systems involved in the elimination of UV-induced damage. Such systems include the immune system, the intracellular repair system, the microcirculation system and not only.
Postreplication repair is the repair of damage to the DNA that takes place after replication.
Transformation efficiency is the efficiency by which cells can take up extracellular DNA and express genes encoded by it. This is based on the competence of the cells. It can be calculated by dividing the number of successful transformants by the amount of DNA used during a transformation procedure. Transformants are cells that have taken up DNA and which can express genes on the introduced DNA.

DNA polymerase eta, is a protein that in humans is encoded by the POLH gene.

Bioreporters are intact, living microbial cells that have been genetically engineered to produce a measurable signal in response to a specific chemical or physical agent in their environment. Bioreporters contain two essential genetic elements, a promoter gene and a reporter gene. The promoter gene is turned on (transcribed) when the target agent is present in the cell’s environment. The promoter gene in a normal bacterial cell is linked to other genes that are then likewise transcribed and then translated into proteins that help the cell in either combating or adapting to the agent to which it has been exposed. In the case of a bioreporter, these genes, or portions thereof, have been removed and replaced with a reporter gene. Consequently, turning on the promoter gene now causes the reporter gene to be turned on. Activation of the reporter gene leads to production of reporter proteins that ultimately generate some type of a detectable signal. Therefore, the presence of a signal indicates that the bioreporter has sensed a particular target agent in its environment.
DNA damage is an alteration in the chemical structure of DNA, such as a break in a strand of DNA, a nucleobase missing from the backbone of DNA, or a chemically changed base such as 8-OHdG. DNA damage can occur naturally or via environmental factors, but is distinctly different from mutation, although both are types of error in DNA. DNA damage is an abnormal chemical structure in DNA, while a mutation is a change in the sequence of base pairs. DNA damages cause changes in the structure of the genetic material and prevents the replication mechanism from functioning and performing properly. The DNA damage response (DDR) is a complex signal transduction pathway which recognizes when DNA is damaged and initiates the cellular response to the damage.
Maxicell is a system for identifying plasmid-encoded proteins that was developed by Sancar, AM Hack, and WD Rupp. The method uses a mutant strain of the bacterium Escherichia coli that is defective in repairing DNA damage. Irradiating the mutant cells with UV light leads to degradation of the bacterial chromosome. Plasmids in the cells mostly escape the UV-induced damage because of their small size. Proteins encoded by genes on the bacterial chromosome will no longer be synthesized, while proteins encoded by genes on the plasmid DNA molecules will continue to be made. The addition of radioactively-labeled amino acids following the UV treatment allows the plasmid-encoded proteins to be specifically visualized, because proteins synthesized prior to the UV treatment will not contain the radioactive label. The method was used for amplification of photolyase in Escherichia coli.













