
In molecular biology, post-translational modification (PTM) is the covalent process of changing proteins following protein biosynthesis. PTMs may involve enzymes or occur spontaneously. Proteins are created by ribosomes, which translate mRNA into polypeptide chains, which may then change to form the mature protein product. PTMs are important components in cell signalling, as for example when prohormones are converted to hormones.
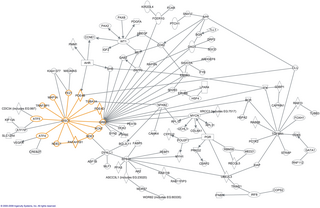
In molecular biology, an interactome is the whole set of molecular interactions in a particular cell. The term specifically refers to physical interactions among molecules but can also describe sets of indirect interactions among genes.
A sequence profiling tool in bioinformatics is a type of software that presents information related to a genetic sequence, gene name, or keyword input. Such tools generally take a query such as a DNA, RNA, or protein sequence or ‘keyword’ and search one or more databases for information related to that sequence. Summaries and aggregate results are provided in standardized format describing the information that would otherwise have required visits to many smaller sites or direct literature searches to compile. Many sequence profiling tools are software portals or gateways that simplify the process of finding information about a query in the large and growing number of bioinformatics databases. The access to these kinds of tools is either web based or locally downloadable executables.
BRENDA is the world's most comprehensive online database for functional, biochemical and molecular biological data on enzymes, metabolites and metabolic pathways. It contains data on the properties, function and significance of all enzymes classified by the Enzyme Commission of the International Union of Biochemistry and Molecular Biology (IUBMB) classified enzymes. As ELIXIR Core Data Resource, BRENDA is considered a data resource of critical importance to the international life sciences research community. The database compiles a representative overview of enzymes and metabolites using current research data from primary scientific literature and thus serves the purpose of facilitating information retrieval for researchers. BRENDA is subject to the terms of the Creative Commons license, is accessible worldwide and can be used free of charge. As one of the digital resources of the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures, BRENDA is part of the integrated biodata infrastructure DSMZ Digital Diversity.

Protein–protein interactions (PPIs) are physical contacts of high specificity established between two or more protein molecules as a result of biochemical events steered by interactions that include electrostatic forces, hydrogen bonding and the hydrophobic effect. Many are physical contacts with molecular associations between chains that occur in a cell or in a living organism in a specific biomolecular context.
InterPro is a database of protein families, protein domains and functional sites in which identifiable features found in known proteins can be applied to new protein sequences in order to functionally characterise them.

The Biological General Repository for Interaction Datasets (BioGRID) is a curated biological database of protein-protein interactions, genetic interactions, chemical interactions, and post-translational modifications created in 2003 (originally referred to as simply the General Repository for Interaction Datasets by Mike Tyers, Bobby-Joe Breitkreutz, and Chris Stark at the Lunenfeld-Tanenbaum Research Institute at Mount Sinai Hospital. It strives to provide a comprehensive curated resource for all major model organism species while attempting to remove redundancy to create a single mapping of data. Users of The BioGRID can search for their protein, chemical or publication of interest and retrieve annotation, as well as curated data as reported, by the primary literature and compiled by in house large-scale curation efforts. The BioGRID is hosted in Toronto, Ontario, Canada and Dallas, Texas, United States and is partnered with the Saccharomyces Genome Database, FlyBase, WormBase, PomBase, and the Alliance of Genome Resources. The BioGRID is funded by the NIH and CIHR. BioGRID is an observer member of the International Molecular Exchange Consortium.
NetPath is a manually curated resource of human signal transduction pathways. It is a joint effort between Pandey Lab at the Johns Hopkins University and the Institute of Bioinformatics (IOB), Bangalore, India, and is also worked on by other parties.
Human Proteinpedia, which is closely associated with Institute of Bioinformatics (IOB), Bangalore and Johns Hopkins University, is a portal for sharing and integration of human proteomic data. It allows research laboratories to contribute and maintain protein annotations. Human Protein Reference Database (HPRD) integrates data, that is deposited in Human Proteinpedia along with the existing literature curated information at the context of an individual protein. In essence, researchers can add new data to HPRD by registering to Human Proteinpedia. The data deposited in Human Proteinpedia is freely available for download. Emphasizing the importance of proteomics data disposition to public repositories, Nature Methods recommends Human Proteinpedia in their editorial. More than 70 labs participate in this effort.

DnaJ homolog subfamily B member 11 is a protein that in humans is encoded by the DNAJB11 gene.

40S ribosomal protein S10 is a protein that in humans is encoded by the RPS10 gene.
The Reference Sequence (RefSeq) database is an open access, annotated and curated collection of publicly available nucleotide sequences and their protein products. RefSeq was introduced in 2000. This database is built by National Center for Biotechnology Information (NCBI), and, unlike GenBank, provides only a single record for each natural biological molecule for major organisms ranging from viruses to bacteria to eukaryotes.
The Eukaryotic Linear Motif (ELM) resource is a computational biology resource for investigating short linear motifs (SLiMs) in eukaryotic proteins. It is currently the largest collection of linear motif classes with annotated and experimentally validated linear motif instances.

In molecular biology short linear motifs (SLiMs), linear motifs or minimotifs are short stretches of protein sequence that mediate protein–protein interaction.
The Death Domain database is a secondary database of protein-protein interactions (PPI) of the death domain superfamily. Members of this superfamily are key players in apoptosis, inflammation, necrosis, and immune cell signaling pathways. Negative death domain superfamily-mediated signaling events result in various human diseases which include, cancers, neurodegenerative diseases, and immunological disorders. Creating death domain databases are of particular interest to researchers in the biomedical field as it enables a further understanding of the molecular mechanisms involved in death domain interactions while also providing easy access to tools such as an interaction map that illustrates the protein-protein interaction network and information. There is currently only one database that exclusively looks at death domains but there are other databases and resources that have information on this superfamily. According to PubMed, this database has been cited by seven peer-reviewed articles to date because of its extensive and specific information on the death domains and their PPI summaries.

Solute carrier family 46 member 3 (SLC46A3) is a protein that in humans is encoded by the SLC46A3 gene. Also referred to as FKSG16, the protein belongs to the major facilitator superfamily (MFS) and SLC46A family. Most commonly found in the plasma membrane and endoplasmic reticulum (ER), SLC46A3 is a multi-pass membrane protein with 11 α-helical transmembrane domains. It is mainly involved in the transport of small molecules across the membrane through the substrate translocation pores featured in the MFS domain. The protein is associated with breast and prostate cancer, hepatocellular carcinoma (HCC), papilloma, glioma, obesity, and SARS-CoV. Based on the differential expression of SLC46A3 in antibody-drug conjugate (ADC)-resistant cells and certain cancer cells, current research is focused on the potential of SLC46A3 as a prognostic biomarker and therapeutic target for cancer. While protein abundance is relatively low in humans, high expression has been detected particularly in the liver, small intestine, and kidney.
The human interactome is the set of protein–protein interactions that occur in human cells. The sequencing of reference genomes, in particular the Human Genome Project, has revolutionized human genetics, molecular biology, and clinical medicine. Genome-wide association study results have led to the association of genes with most Mendelian disorders, and over 140 000 germline mutations have been associated with at least one genetic disease. However, it became apparent that inherent to these studies is an emphasis on clinical outcome rather than a comprehensive understanding of human disease; indeed to date the most significant contributions of GWAS have been restricted to the “low-hanging fruit” of direct single mutation disorders, prompting a systems biology approach to genomic analysis. The connection between genotype and phenotype remain elusive, especially in the context of multigenic complex traits and cancer. To assign functional context to genotypic changes, much of recent research efforts have been devoted to the mapping of the networks formed by interactions of cellular and genetic components in humans, as well as how these networks are altered by genetic and somatic disease.

CCDC188 or coiled-coil domain containing protein is a protein that in humans is encoded by the CCDC188 gene.
The Institute of Bioinformatics, often referred to as IOB, is an Indian not-for-profit academic research organization based in Bangalore, India. It is involved in research in the fields of bioinformatics, multi-omics, systems biology and neurological disorders. In 2002, the institute was set up by The Genomics Research Trust and the Johns Hopkins University of Baltimore, Maryland. This organization is recognized as a 'Scientific and Industrial Research Organization' (SIRO) of the Department of Scientific and Industrial Research, Government of India. Renowned Proteomicist Akhilesh Pandey, Professor at Department of Laboratory Medicine and Pathology, Center for Individualized Medicine of Mayo Clinic in Rochester, Minnesota, USA is the Founding and current Director of IOB, and eminent Proteomicist Ravi Sirdeshmukh, Founder President of the 'Proteomic Society of India' is the current Associate Director of IOB.









