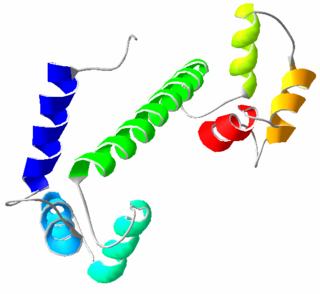
Calmodulin (CaM) (an abbreviation for calcium-modulated protein) is a multifunctional intermediate calcium-binding messenger protein expressed in all eukaryotic cells. It is an intracellular target of the secondary messenger Ca2+, and the binding of Ca2+ is required for the activation of calmodulin. Once bound to Ca2+, calmodulin acts as part of a calcium signal transduction pathway by modifying its interactions with various target proteins such as kinases or phosphatases.
In biology, a sequence motif is a nucleotide or amino-acid sequence pattern that is widespread and usually assumed to be related to biological function of the macromolecule. For example, an N-glycosylation site motif can be defined as Asn, followed by anything but Pro, followed by either Ser or Thr, followed by anything but Pro residue.
Myosins are a superfamily of motor proteins best known for their roles in muscle contraction and in a wide range of other motility processes in eukaryotes. They are ATP-dependent and responsible for actin-based motility.

Helix-turn-helix is a DNA-binding protein (DBP). The helix-turn-helix (HTH) is a major structural motif capable of binding DNA. Each monomer incorporates two α helices, joined by a short strand of amino acids, that bind to the major groove of DNA. The HTH motif occurs in many proteins that regulate gene expression. It should not be confused with the helix–loop–helix motif.

Telokin is an abundant protein found in smooth-muscle. It is identical to the C-terminus of myosin light-chain kinase. Telokin may play a role in the stabilization of unphosphorylated smooth-muscle myosin filaments. Because of its origin as the C-terminal end of smooth muscle myosin light chain kinase, it is called "telokin".

Myosin light-chain kinase also known as MYLK or MLCK is a serine/threonine-specific protein kinase that phosphorylates a specific myosin light chain, namely, the regulatory light chain of myosin II.
Calmodulin-binding proteins are, as their name implies, proteins which bind calmodulin. Calmodulin can bind to a variety of proteins through a two-step binding mechanism, namely "conformational and mutually induced fit", where typically two domains of calmodulin wrap around an emerging helical calmodulin binding domain from the target protein.

The EF hand is a helix–loop–helix structural domain or motif found in a large family of calcium-binding proteins.
Phosphodiesterase 1, PDE1, EC 3.1.4.1, systematic name oligonucleotide 5′-nucleotidohydrolase) is a phosphodiesterase enzyme also known as calcium- and calmodulin-dependent phosphodiesterase. It is one of the 11 families of phosphodiesterase (PDE1-PDE11). Phosphodiesterase 1 has three subtypes, PDE1A, PDE1B and PDE1C which divide further into various isoforms. The various isoforms exhibit different affinities for cAMP and cGMP.

Myosin light-chain phosphatase, also called myosin phosphatase (EC 3.1.3.53; systematic name [myosin-light-chain]-phosphate phosphohydrolase), is an enzyme (specifically a serine/threonine-specific protein phosphatase) that dephosphorylates the regulatory light chain of myosin II:

A myosin light chain is a light chain of myosin. Myosin light chains were discovered by Chinese biochemist Cao Tianqin when he was a graduate student at the University of Cambridge in England.

Calcium/calmodulin-dependent protein kinase type II gamma chain is an enzyme that in humans is encoded by the CAMK2G gene.

Myosin essential light chain (ELC), ventricular/cardiac isoform is a protein that in humans is encoded by the MYL3 gene. This cardiac ventricular/slow skeletal ELC isoform is distinct from that expressed in fast skeletal muscle (MYL1) and cardiac atrial muscle (MYL4). Ventricular ELC is part of the myosin molecule and is important in modulating cardiac muscle contraction.

Calcium binding protein 1 is a protein that in humans is encoded by the CABP1 gene. Calcium-binding protein 1 is a calcium-binding protein discovered in 1999. It has two EF hand motifs and is expressed in neuronal cells in such areas as hippocampus, habenular nucleus of the epithalamus, Purkinje cell layer of the cerebellum, and the amacrine cells and cone bipolar cells of the retina.

Myosin X, also known as MYO10, is a protein that in humans is encoded by the MYO10 gene.

Serine/threonine protein kinase WNK4 also known as WNK lysine deficient protein kinase 4 or WNK4, is an enzyme that in humans is encoded by the WNK4 gene. Missense mutations cause a genetic form of pseudohypoaldosteronism type 2, also called Gordon syndrome.

Myosin light chain kinase 2 also known as MYLK2 is an enzyme which in humans is encoded by the MYLK2 gene.
The Walker A and Walker B motifs are protein sequence motifs, known to have highly conserved three-dimensional structures. These were first reported in ATP-binding proteins by Walker and co-workers in 1982.

The WRKY domain is found in the WRKY transcription factor family, a class of transcription factors. The WRKY domain is found almost exclusively in plants although WRKY genes appear present in some diplomonads, social amoebae and other amoebozoa, and fungi incertae sedis. They appear absent in other non-plant species. WRKY transcription factors have been a significant area of plant research for the past 20 years. The WRKY DNA-binding domain recognizes the W-box (T)TGAC(C/T) cis-regulatory element.

LRRIQ3, which is also known as LRRC44, is a protein that in humans is encoded by the LRRIQ3 gene. It is predominantly expressed in the testes, and is linked to a number of diseases.















