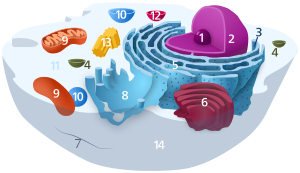
The cell nucleus is a membrane-bound organelle found in eukaryotic cells. Eukaryotic cells usually have a single nucleus, but a few cell types, such as mammalian red blood cells, have no nuclei, and a few others including osteoclasts have many. The main structures making up the nucleus are the nuclear envelope, a double membrane that encloses the entire organelle and isolates its contents from the cellular cytoplasm; and the nuclear matrix, a network within the nucleus that adds mechanical support.

RNA splicing is a process in molecular biology where a newly-made precursor messenger RNA (pre-mRNA) transcript is transformed into a mature messenger RNA (mRNA). It works by removing all the introns and splicing back together exons. For nuclear-encoded genes, splicing occurs in the nucleus either during or immediately after transcription. For those eukaryotic genes that contain introns, splicing is usually needed to create an mRNA molecule that can be translated into protein. For many eukaryotic introns, splicing occurs in a series of reactions which are catalyzed by the spliceosome, a complex of small nuclear ribonucleoproteins (snRNPs). There exist self-splicing introns, that is, ribozymes that can catalyze their own excision from their parent RNA molecule. The process of transcription, splicing and translation is called gene expression, the central dogma of molecular biology.

A spliceosome is a large ribonucleoprotein (RNP) complex found primarily within the nucleus of eukaryotic cells. The spliceosome is assembled from small nuclear RNAs (snRNA) and numerous proteins. Small nuclear RNA (snRNA) molecules bind to specific proteins to form a small nuclear ribonucleoprotein complex, which in turn combines with other snRNPs to form a large ribonucleoprotein complex called a spliceosome. The spliceosome removes introns from a transcribed pre-mRNA, a type of primary transcript. This process is generally referred to as splicing. An analogy is a film editor, who selectively cuts out irrelevant or incorrect material from the initial film and sends the cleaned-up version to the director for the final cut.

Cajal bodies (CBs), also coiled bodies, are spherical nuclear bodies of 0.3–1.0 μm in diameter found in the nucleus of proliferative cells like embryonic cells and tumor cells, or metabolically active cells like neurons. CBs are membrane-less organelles and largely consist of proteins and RNA. They were first reported by Santiago Ramón y Cajal in 1903, who called them nucleolar accessory bodies due to their association with the nucleoli in neuronal cells. They were rediscovered with the use of the electron microscope (EM) and named coiled bodies, according to their appearance as coiled threads on EM images, and later renamed after their discoverer. Research on CBs was accelerated after discovery and cloning of the marker protein p80/Coilin. CBs have been implicated in RNA-related metabolic processes such as the biogenesis, maturation and recycling of snRNPs, histone mRNA processing and telomere maintenance. CBs assemble RNA which is used by telomerase to add nucleotides to the ends of telomeres.

SR proteins are a conserved family of proteins involved in RNA splicing. SR proteins are named because they contain a protein domain with long repeats of serine and arginine amino acid residues, whose standard abbreviations are "S" and "R" respectively. SR proteins are ~200-600 amino acids in length and composed of two domains, the RNA recognition motif (RRM) region and the RS domain. SR proteins are more commonly found in the nucleus than the cytoplasm, but several SR proteins are known to shuttle between the nucleus and the cytoplasm.

A primary transcript is the single-stranded ribonucleic acid (RNA) product synthesized by transcription of DNA, and processed to yield various mature RNA products such as mRNAs, tRNAs, and rRNAs. The primary transcripts designated to be mRNAs are modified in preparation for translation. For example, a precursor mRNA (pre-mRNA) is a type of primary transcript that becomes a messenger RNA (mRNA) after processing.
snRNPs, or small nuclear ribonucleoproteins, are RNA-protein complexes that combine with unmodified pre-mRNA and various other proteins to form a spliceosome, a large RNA-protein molecular complex upon which splicing of pre-mRNA occurs. The action of snRNPs is essential to the removal of introns from pre-mRNA, a critical aspect of post-transcriptional modification of RNA, occurring only in the nucleus of eukaryotic cells. Additionally, U7 snRNP is not involved in splicing at all, as U7 snRNP is responsible for processing the 3′ stem-loop of histone pre-mRNA.
Small nuclear RNA (snRNA) is a class of small RNA molecules that are found within the splicing speckles and Cajal bodies of the cell nucleus in eukaryotic cells. The length of an average snRNA is approximately 150 nucleotides. They are transcribed by either RNA polymerase II or RNA polymerase III. Their primary function is in the processing of pre-messenger RNA (hnRNA) in the nucleus. They have also been shown to aid in the regulation of transcription factors or RNA polymerase II, and maintaining the telomeres.

Heterogeneous nuclear ribonucleoproteins (hnRNPs) are complexes of RNA and protein present in the cell nucleus during gene transcription and subsequent post-transcriptional modification of the newly synthesized RNA (pre-mRNA). The presence of the proteins bound to a pre-mRNA molecule serves as a signal that the pre-mRNA is not yet fully processed and therefore not ready for export to the cytoplasm. Since most mature RNA is exported from the nucleus relatively quickly, most RNA-binding protein in the nucleus exist as heterogeneous ribonucleoprotein particles. After splicing has occurred, the proteins remain bound to spliced introns and target them for degradation.

U1 spliceosomal RNA is the small nuclear RNA (snRNA) component of U1 snRNP, an RNA-protein complex that combines with other snRNPs, unmodified pre-mRNA, and various other proteins to assemble a spliceosome, a large RNA-protein molecular complex upon which splicing of pre-mRNA occurs. Splicing, or the removal of introns, is a major aspect of post-transcriptional modification, and takes place only in the nucleus of eukaryotes.

Nuclear bodies are biomolecular condensates, membraneless structures found in the cell nuclei of eukaryotic cells. Nuclear bodies include Cajal bodies, the nucleolus, nuclear speckles, histone locus bodies, and promyelocytic leukemia protein (PML) nuclear bodies. Nuclear bodies also include ND10s. ND stands for nuclear domain, and 10 refers to the number of dots seen. Additionally, a nuclear body subtype is a clastosome suggested to be a site of protein degradation.

Heterogeneous nuclear ribonucleoprotein K is a protein that in humans is encoded by the HNRNPK gene. It is found in the cell nucleus that binds to pre-messenger RNA (mRNA) as a component of heterogeneous ribonucleoprotein particles. The simian homolog is known as protein H16. Both proteins bind to single-stranded DNA as well as to RNA and can stimulate the activity of RNA polymerase II, the protein responsible for most gene transcription. The relative affinities of the proteins for DNA and RNA vary with solution conditions and are inversely correlated, so that conditions promoting strong DNA binding result in weak RNA binding.

Splicing factor U2AF 65 kDa subunit is a protein that in humans is encoded by the U2AF2 gene.

snRNP70 also known as U1 small nuclear ribonucleoprotein 70 kDa is a protein that in humans is encoded by the SNRNP70 gene. snRNP70 is a small nuclear ribonucleoprotein that associates with U1 spliceosomal RNA, forming the U1snRNP a core component of the spliceosome. The U1-70K protein and other components of the spliceosome complex form detergent-insoluble aggregates in both sporadic and familial human cases of Alzheimer's disease. U1-70K co-localizes with Tau in neurofibrillary tangles in Alzheimer's disease.
U4/U6 small nuclear ribonucleoprotein Prp3 is a protein that in humans is encoded by the PRPF3 gene.

Polypyrimidine tract-binding protein 1 is a protein that in humans is encoded by the PTBP1 gene.

Serine/arginine-rich splicing factor 1 (SRSF1) also known as alternative splicing factor 1 (ASF1), pre-mRNA-splicing factor SF2 (SF2) or ASF1/SF2 is a protein that in humans is encoded by the SRSF1 gene. ASF/SF2 is an essential sequence specific splicing factor involved in pre-mRNA splicing. SRSF1 is the gene that codes for ASF/SF2 and is found on chromosome 17. The resulting splicing factor is a protein of approximately 33 kDa. ASF/SF2 is necessary for all splicing reactions to occur, and influences splice site selection in a concentration-dependent manner, resulting in alternative splicing. In addition to being involved in the splicing process, ASF/SF2 also mediates post-splicing activities, such as mRNA nuclear export and translation.

Prp24 is a protein part of the pre-messenger RNA splicing process and aids the binding of U6 snRNA to U4 snRNA during the formation of spliceosomes. Found in eukaryotes from yeast to E. coli, fungi, and humans, Prp24 was initially discovered to be an important element of RNA splicing in 1989. Mutations in Prp24 were later discovered in 1991 to suppress mutations in U4 that resulted in cold-sensitive strains of yeast, indicating its involvement in the reformation of the U4/U6 duplex after the catalytic steps of splicing.
Messenger RNP is mRNA with bound proteins. mRNA does not exist "naked" in vivo but is always bound by various proteins while being synthesized, spliced, exported, and translated in the cytoplasm.
Snurposomes are a type of granular structure that are found in the nuclei of oocytes. They are found in animals including mammals, amphibians, and insects. Snurposomes’ role in the cell is to contain small nuclear ribonucleoproteins, otherwise known as snRNPs.