Related Research Articles

Saccharomyces cerevisiae is a species of yeast. The species has been instrumental in winemaking, baking, and brewing since ancient times. It is believed to have been originally isolated from the skin of grapes. It is one of the most intensively studied eukaryotic model organisms in molecular and cell biology, much like Escherichia coli as the model bacterium. It is the microorganism which causes many common types of fermentation. S. cerevisiae cells are round to ovoid, 5–10 μm in diameter. It reproduces by budding.

Histone methyltransferases (HMT) are histone-modifying enzymes, that catalyze the transfer of one, two, or three methyl groups to lysine and arginine residues of histone proteins. The attachment of methyl groups occurs predominantly at specific lysine or arginine residues on histones H3 and H4. Two major types of histone methyltranferases exist, lysine-specific and arginine-specific. In both types of histone methyltransferases, S-Adenosyl methionine (SAM) serves as a cofactor and methyl donor group.
The genomic DNA of eukaryotes associates with histones to form chromatin. The level of chromatin compaction depends heavily on histone methylation and other post-translational modifications of histones. Histone methylation is a principal epigenetic modification of chromatin that determines gene expression, genomic stability, stem cell maturation, cell lineage development, genetic imprinting, DNA methylation, and cell mitosis.

Non-homologous end joining (NHEJ) is a pathway that repairs double-strand breaks in DNA. It is called "non-homologous" because the break ends are directly ligated without the need for a homologous template, in contrast to homology directed repair (HDR), which requires a homologous sequence to guide repair. NHEJ is active in both non-dividing and proliferating cells, while HDR is not readily accessible in non-dividing cells. The term "non-homologous end joining" was coined in 1996 by Moore and Haber.

DNA replication licensing factor MCM6 is a protein that in humans is encoded by the MCM6 gene. MCM6 is one of the highly conserved mini-chromosome maintenance proteins (MCM) that are essential for the initiation of eukaryotic genome replication.
Mitotic recombination is a type of genetic recombination that may occur in somatic cells during their preparation for mitosis in both sexual and asexual organisms. In asexual organisms, the study of mitotic recombination is one way to understand genetic linkage because it is the only source of recombination within an individual. Additionally, mitotic recombination can result in the expression of recessive alleles in an otherwise heterozygous individual. This expression has important implications for the study of tumorigenesis and lethal recessive alleles. Mitotic homologous recombination occurs mainly between sister chromatids subsequent to replication. Inter-sister homologous recombination is ordinarily genetically silent. During mitosis the incidence of recombination between non-sister homologous chromatids is only about 1% of that between sister chromatids.
The mating-type locus is a specialized region in the genomes of some yeast and other fungi, usually organized into heterochromatin and possessing unique histone methylation patterns. The genes in this region regulate the mating type of the organism and therefore determine key events in its life cycle, such as whether it will reproduce sexually or asexually. In fission yeast such as S. pombe, the formation and maintenance of the heterochromatin organization is regulated by RNA-induced transcriptional silencing, a form of RNA interference responsible for genomic maintenance in many organisms. Mating type regions have also been well studied in budding yeast S. cerevisiae and in the fungus Neurospora crassa.

DNA replication licensing factor MCM2 is a protein that in humans is encoded by the MCM2 gene.

UV excision repair protein RAD23 homolog A is a protein that in humans is encoded by the RAD23A gene.

DNA replication licensing factor MCM4 is a protein that in humans is encoded by the MCM4 gene.

DNA repair protein RAD50, also known as RAD50, is a protein that in humans is encoded by the RAD50 gene.

Exonuclease 1 is an enzyme that in humans is encoded by the EXO1 gene.

RAD52 homolog , also known as RAD52, is a protein which in humans is encoded by the RAD52 gene.

DNA repair protein RAD51 homolog 4 is a protein that in humans is encoded by the RAD51L3 gene.
SAE2 is a gene in budding yeast, coding for the protein Sae2, which is involved in DNA repair. Sae2 is a part of the homologous recombination process in response to double-strand breaks. It is best characterized in the yeast model organism Saccharomyces cerevisiae. Homologous genes in other organisms include Ctp1 in fission yeast, Com1 in plants, and CtIP in higher eukaryotes including humans.
Protein HIRA is a protein that in humans is encoded by the HIRA gene. This gene is mapped to 22q11.21, centromeric to COMT.

DnaJ homolog subfamily A member 2 is a protein that in humans is encoded by the DNAJA2 gene.

Structural maintenance of chromosomes protein 6 is a protein that in humans is encoded by the SMC6 gene.

Nucleosome assembly protein 1-like 4 is a protein that in humans is encoded by the NAP1L4 gene.
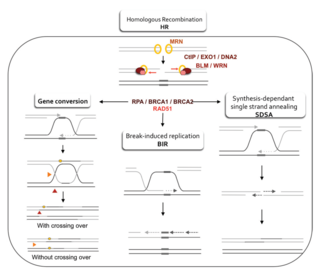
Homology-directed repair (HDR) is a mechanism in cells to repair double-strand DNA lesions. The most common form of HDR is homologous recombination. The HDR mechanism can only be used by the cell when there is a homologous piece of DNA present in the nucleus, mostly in G2 and S phase of the cell cycle. Other examples of homology-directed repair include single-strand annealing and breakage-induced replication. When the homologous DNA is absent, another process called non-homologous end joining (NHEJ) takes place instead.
SilentInformationRegulator (SIR) proteins are involved in regulating gene expression. SIR proteins organize heterochromatin near telomeres, ribosomal DNA (rDNA), and at silent loci including hidden mating type loci in yeast. The SIR family of genes encodes catalytic and non-catalytic proteins that are involved in de-acetylation of histone tails and the subsequent condensation of chromatin around a SIR protein scaffold. Some SIR family members are conserved from yeast to humans.
References
- 1 2 3 4 5 "James Haber". www.nasonline.org.
- ↑ Haber, J. E.; Koshland, D. E. Jr. (1967). "Relation of protein subunit interactions to the molecular species observed during cooperative binding of ligands". Proc. Natl. Acad. Sci. USA. 58 (5): 2087–2093. Bibcode:1967PNAS...58.2087H. doi: 10.1073/pnas.58.5.2087 . PMC 223909 . PMID 5237502.
- ↑ Haber, J. E.; Koshland, D. E. Jr. (1969). "Evidence for β-β interactions during the binding of oxygen to hemoglobin". Biochim. Biophys. Acta. 194 (1): 339–341. doi:10.1016/0005-2795(69)90215-3. PMID 5353133.
- ↑ Haber, J. E.; Koshland, D. E. Jr. (1970). "An evaluation of the relatedness of proteins based on comparison of amino acid sequences". J. Mol. Biol. 50 (3): 617–639. doi:10.1016/0022-2836(70)90089-6. PMID 4097749.
- ↑ Haber, J. E.; Halvorson, H.O. (1972). "Cell cycle dependency of sporulation in Saccharomyces cerevisiae". J. Bacteriol. 109 (3): 1027–1033. doi:10.1128/JB.109.3.1027-1033.1972. PMC 247323 . PMID 4551739.
- ↑ Haber, J. E. (2000). "Partners and pathways: Repairing a double-strand break". Trends Genet. 16 (6): 259–264. doi:10.1016/S0168-9525(00)02022-9. PMID 10827453.
- ↑ Harrison, J. C.; Haber, J. E. (2006). "Surviving the breakup: The DNA damage checkpoint". Annu. Rev. Genet. 40: 209–235. doi:10.1146/annurev.genet.40.051206.105231. PMID 16805667.
- ↑ Moore, J. K.; Haber, J. E. (1996). "Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae". Mol. Cell. Biol. 16 (5): 2164–2173. doi:10.1128/mcb.16.5.2164. PMC 231204 . PMID 8628283.
- ↑ De Koning, L.; Corpet, A.; Haber, J. E.; Almouzni, G. (2007). "Histone chaperones: an escort network regulating histone traffic". Nat. Struct. Mol. Biol. 14 (11): 997–1007. doi:10.1038/nsmb1318. PMID 17984962. S2CID 38172713.