Related Research Articles

Bioinformatics is an interdisciplinary field that develops methods and software tools for understanding biological data, in particular when the data sets are large and complex. As an interdisciplinary field of science, bioinformatics combines biology, chemistry, physics, computer science, information engineering, mathematics and statistics to analyze and interpret the biological data. Bioinformatics has been used for in silico analyses of biological queries using computational and statistical techniques.

In the fields of molecular biology and genetics, a genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA. The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as regulatory sequences, and often a substantial fraction of junk DNA with no evident function. Almost all eukaryotes have mitochondria and a small mitochondrial genome. Algae and plants also contain chloroplasts with a chloroplast genome.

The human genome is a complete set of nucleic acid sequences for humans, encoded as DNA within the 23 chromosome pairs in cell nuclei and in a small DNA molecule found within individual mitochondria. These are usually treated separately as the nuclear genome and the mitochondrial genome. Human genomes include both protein-coding DNA sequences and various types of DNA that does not encode proteins. The latter is a diverse category that includes DNA coding for non-translated RNA, such as that for ribosomal RNA, transfer RNA, ribozymes, small nuclear RNAs, and several types of regulatory RNAs. It also includes promoters and their associated gene-regulatory elements, DNA playing structural and replicatory roles, such as scaffolding regions, telomeres, centromeres, and origins of replication, plus large numbers of transposable elements, inserted viral DNA, non-functional pseudogenes and simple, highly-repetitive sequences. Introns make up a large percentage of non-coding DNA. Some of this non-coding DNA is non-functional junk DNA, such as pseudogenes, but there is no firm consensus on the total amount of junk DNA.

Genomics is an interdisciplinary field of biology focusing on the structure, function, evolution, mapping, and editing of genomes. A genome is an organism's complete set of DNA, including all of its genes as well as its hierarchical, three-dimensional structural configuration. In contrast to genetics, which refers to the study of individual genes and their roles in inheritance, genomics aims at the collective characterization and quantification of all of an organism's genes, their interrelations and influence on the organism. Genes may direct the production of proteins with the assistance of enzymes and messenger molecules. In turn, proteins make up body structures such as organs and tissues as well as control chemical reactions and carry signals between cells. Genomics also involves the sequencing and analysis of genomes through uses of high throughput DNA sequencing and bioinformatics to assemble and analyze the function and structure of entire genomes. Advances in genomics have triggered a revolution in discovery-based research and systems biology to facilitate understanding of even the most complex biological systems such as the brain.

Genome projects are scientific endeavours that ultimately aim to determine the complete genome sequence of an organism and to annotate protein-coding genes and other important genome-encoded features. The genome sequence of an organism includes the collective DNA sequences of each chromosome in the organism. For a bacterium containing a single chromosome, a genome project will aim to map the sequence of that chromosome. For the human species, whose genome includes 22 pairs of autosomes and 2 sex chromosomes, a complete genome sequence will involve 46 separate chromosome sequences.
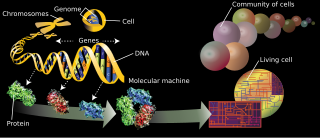
The branches of science known informally as omics are various disciplines in biology whose names end in the suffix -omics, such as genomics, proteomics, metabolomics, metagenomics, phenomics and transcriptomics. Omics aims at the collective characterization and quantification of pools of biological molecules that translate into the structure, function, and dynamics of an organism or organisms.
A genomic library is a collection of overlapping DNA fragments that together make up the total genomic DNA of a single organism. The DNA is stored in a population of identical vectors, each containing a different insert of DNA. In order to construct a genomic library, the organism's DNA is extracted from cells and then digested with a restriction enzyme to cut the DNA into fragments of a specific size. The fragments are then inserted into the vector using DNA ligase. Next, the vector DNA can be taken up by a host organism - commonly a population of Escherichia coli or yeast - with each cell containing only one vector molecule. Using a host cell to carry the vector allows for easy amplification and retrieval of specific clones from the library for analysis.

The Human Genome Project (HGP) was an international scientific research project with the goal of determining the base pairs that make up human DNA, and of identifying, mapping and sequencing all of the genes of the human genome from both a physical and a functional standpoint. It started in 1990 and was completed in 2003. It remains the world's largest collaborative biological project. Planning for the project started after it was adopted in 1984 by the US government, and it officially launched in 1990. It was declared complete on April 14, 2003, and included about 92% of the genome. Level "complete genome" was achieved in May 2021, with a remaining only 0.3% bases covered by potential issues. The final gapless assembly was finished in January 2022.

Olfactory receptor 10H2 is a protein that in humans is encoded by the OR10H2 gene.

Olfactory receptor 10H3 is a protein that in humans is encoded by the OR10H3 gene.

Olfactory receptor 7A5 is a protein that in humans is encoded by the OR7A5 gene.

Kallikrein-12 is a protein that in humans is encoded by the KLK12 gene.

Olfactory receptor 1I1 is a protein that in humans is encoded by the OR1I1 gene.

Olfactory receptor 7A10 is a protein that in humans is encoded by the OR7A10 gene.

Endothelial zinc finger protein induced by tumor necrosis factor alpha is a protein that in humans is encoded by the ZNF71 gene.

Zinc finger protein 264 is a protein that in humans is encoded by the ZNF264 gene.

A reference genome is a digital nucleic acid sequence database, assembled by scientists as a representative example of the set of genes in one idealized individual organism of a species. As they are assembled from the sequencing of DNA from a number of individual donors, reference genomes do not accurately represent the set of genes of any single individual organism. Instead a reference provides a haploid mosaic of different DNA sequences from each donor. For example, the most recent human reference genome is derived from >60 genomic clone libraries. There are reference genomes for multiple species of viruses, bacteria, fungus, plants, and animals. Reference genomes are typically used as a guide on which new genomes are built, enabling them to be assembled much more quickly and cheaply than the initial Human Genome Project. Reference genomes can be accessed online at several locations, using dedicated browsers such as Ensembl or UCSC Genome Browser.

George M. Weinstock is an American geneticist and microbiologist on the faculty of The Jackson Laboratory for Genomic Medicine, where he is a professor and the associate director for microbial genomics. Before joining The Jackson Laboratory, he taught at Washington University in St. Louis and served as associate director of The Genome Institute. Previously, Dr. Weinstock was co-director of the Human Genome Sequencing Center (HGSC) at Baylor College of Medicine in Houston, Texas, and Professor of Molecular and Human Genetics there.[1] He received his B.S. degree from the University of Michigan in 1970 and his Ph.D. from the Massachusetts Institute of Technology in 1977. He has spent most of his career taking genomic approaches to study fundamental biological processes.
Catherine Feuillet is a French geneticist who is currently the Chief Scientific Officer of Inari Agriculture, a Cambridge MA based biotechnology company. Feuillet earned a PhD in plant molecular biology on the isolation and characterization of genes involved in wood formation in eucalyptus trees. She started to work on the genetics of disease resistance in wheat in 1994 during her post-doctoral studies at the Swiss Federal Institute for Agroecology. She then moved as a junior group leader to the University of Zurich where she investigated the molecular basis of fungal disease resistance in wheat and in barley and cloned the first leaf rust resistance gene from wheat. In 2004 she was hired as a research director at the Institut National de la Recherche Agronomique (INRA) in France to lead European and international projects on wheat genomics.
References
- 1 2 3 4 "Get to Know: Jane Grimwood". hudsonalpha.org. Archived from the original on 23 November 2015. Retrieved 2 November 2015.
- 1 2 3 "Jane Grimwood". hagsc.org. Archived from the original on 26 November 2015. Retrieved 2 November 2015.
- ↑ "GNN - Two More Human Chromosomes Are Complete". www.genomenewsnetwork.org. Retrieved 2017-03-02.
- ↑ Grimwood, Jane; Gordon, Laurie A.; Olsen, Anne; Terry, Astrid; Schmutz, Jeremy; Lamerdin, Jane; Hellsten, Uffe; Goodstein, David; Couronne, Olivier (2004-04-01). "The DNA sequence and biology of human chromosome 19". Nature. 428 (6982): 529–535. Bibcode:2004Natur.428..529G. doi: 10.1038/nature02399 . ISSN 1476-4687. PMID 15057824.
- ↑ JIM MELVIN Clemson University. "COTTON 'MAP': Clemson scientist shares $2.4 million from NSF to advance genomic research". The Times and Democrat. Retrieved 2 November 2015.
- ↑ Liz Hurley (27 July 2015). "HudsonAlpha investigator, research partners receive $2M to seque - WAFF-TV: News, Weather and Sports for Huntsville, AL". waff.com. Retrieved 2 November 2015.
- ↑ "Clemson scientist shares $2.4 million from NSF to advance cotton genomic research". clemson.edu. Retrieved 2 November 2015.