Transposase is an enzyme that binds to the end of a transposon and catalyzes its movement to another part of the genome by a cut and paste mechanism or a replicative transposition mechanism. The word "transposase" was first coined by the individuals who cloned the enzyme required for transposition of the Tn3 transposon. The existence of transposons was postulated in the late 1940s by Barbara McClintock, who was studying the inheritance of maize, but the actual molecular basis for transposition was described by later groups. McClintock discovered that pieces of the chromosomes changed their position, jumping from one chromosome to another. The repositioning of these transposons allowed other genes for pigment to be expressed. Transposition in maize causes changes in color; however, in other organisms, such as bacteria, it can cause antibiotic resistance. Transposition is also important in creating genetic diversity within species and adaptability to changing living conditions. During the course of human evolution, as much as 40% of the human genome has moved around via methods such as transposition of transposons.

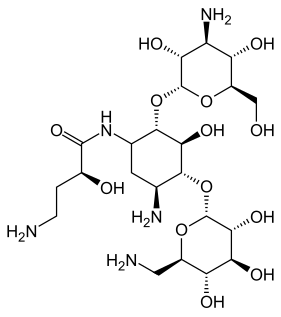
Kanamycin A, often referred to simply as kanamycin, is an antibiotic used to treat severe bacterial infections and tuberculosis. It is not a first line treatment. It is used by mouth, injection into a vein, or injection into a muscle. Kanamycin is recommended for short-term use only, usually from 7 to 10 days. As with most antibiotics, it is ineffective in viral infections.

Pyruvate decarboxylase is a homotetrameric enzyme that catalyses the decarboxylation of pyruvic acid to acetaldehyde and carbon dioxide in the cytoplasm of prokaryotes, and in the cytoplasm and mitochondria of eukaryotes. It is also called 2-oxo-acid carboxylase, alpha-ketoacid carboxylase, and pyruvic decarboxylase. In anaerobic conditions, this enzyme is part of the fermentation process that occurs in yeast, especially of the genus Saccharomyces, to produce ethanol by fermentation. It is also present in some species of fish where it permits the fish to perform ethanol fermentation when oxygen is scarce. Pyruvate decarboxylase starts this process by converting pyruvate into acetaldehyde and carbon dioxide. Pyruvate decarboxylase depends on cofactors thiamine pyrophosphate (TPP) and magnesium. This enzyme should not be mistaken for the unrelated enzyme pyruvate dehydrogenase, an oxidoreductase, that catalyzes the oxidative decarboxylation of pyruvate to acetyl-CoA.
Amikacin is an antibiotic used for a number of bacterial infections. This includes joint infections, intra-abdominal infections, meningitis, pneumonia, sepsis, and urinary tract infections. It is also used for the treatment of multidrug-resistant tuberculosis. It is used either by injection into a vein or muscle.
A selectable marker is a gene introduced into a cell, especially a bacterium or to cells in culture, that confers a trait suitable for artificial selection. They are a type of reporter gene used in laboratory microbiology, molecular biology, and genetic engineering to indicate the success of a transfection or other procedure meant to introduce foreign DNA into a cell. Selectable markers are often antibiotic resistance genes. Bacteria that have been subjected to a procedure to introduce foreign DNA are grown on a medium containing an antibiotic, and those bacterial colonies that can grow have successfully taken up and expressed the introduced genetic material. Normally the genes encoding resistance to antibiotics such as ampicillin, chloroamphenicol, tetracycline or kanamycin, etc., are considered useful selectable markers for E. coli.
Penicillin-binding proteins (PBPs) are a group of proteins that are characterized by their affinity for and binding of penicillin. They are a normal constituent of many bacteria; the name just reflects the way by which the protein was discovered. All β-lactam antibiotics bind to PBPs, which are essential for bacterial cell wall synthesis. PBPs are members of a subgroup of enzymes called transpeptidases. Specifically, PBPs are DD-transpeptidases.
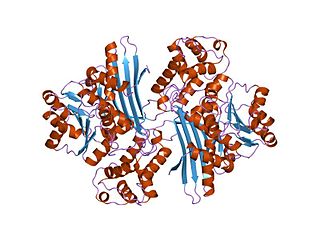
In molecular biology, the protein domain Saccharopine dehydrogenase (SDH), also named Saccharopine reductase, is an enzyme involved in the metabolism of the amino acid lysine, via an intermediate substance called saccharopine. The Saccharopine dehydrogenase enzyme can be classified under EC 1.5.1.7, EC 1.5.1.8, EC 1.5.1.9, and EC 1.5.1.10. It has an important function in lysine metabolism and catalyses a reaction in the alpha-Aminoadipic acid pathway. This pathway is unique to fungal organisms therefore, this molecule could be useful in the search for new antibiotics. This protein family also includes saccharopine dehydrogenase and homospermidine synthase. It is found in prokaryotes, eukaryotes and archaea.

Nucleotidyltransferases are transferase enzymes of phosphorus-containing groups, e.g., substituents of nucleotidylic acids or simply nucleoside monophosphates. The general reaction of transferring a nucleoside monophosphate moiety from A to B, can be written as:

RNase PH is a 3'-5' exoribonuclease and nucleotidyltransferase, present in archaea and bacteria, that is involved in tRNA processing. Contrary to hydrolytic enzymes, it is a phosphorolytic enzyme, meaning that it uses inorganic phosphate as a reactant to cleave nucleotide-nucleotide bonds, releasing diphosphate nucleotides. The active structure of the proteins is a homohexameric complex, consisting of three ribonuclease (RNase) PH dimers. RNase PH has homologues in many other organisms, which are referred to as RNase PH-like proteins. The part of another larger protein with a domain that is very similar to RNase PH is called an RNase PH domain (RPD).
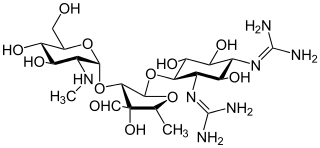
In enzymology, an aminoglycoside N6'-acetyltransferase (EC 2.3.1.82) is an enzyme that catalyzes the chemical reaction
In enzymology, an aldose-1-phosphate nucleotidyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a galactose-1-phosphate thymidylyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a gentamicin 2"-nucleotidyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a glucuronate-1-phosphate uridylyltransferase is an enzyme that catalyzes the chemical reaction
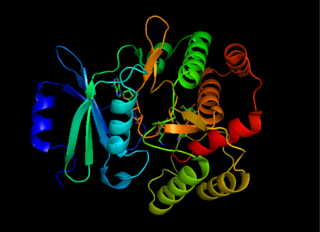
Aminoglycoside-3'-phosphotransferase, also known as aminoglycoside kinase, is an enzyme that primarily catalyzes the addition of phosphate from ATP to the 3'-hydroxyl group of a 4,6-disubstituted aminoglycoside, such as kanamycin. However, APH(3') has also been found to phosphorylate at the 5'-hydroxyl group in 4,5-disubstituted aminoglycosides, which lack a 3'-hydroxyl group, and to diphosphorylate hydroxyl groups in aminoglycosides that have both 3'- and 5'-hydroxyl groups. Primarily positively charged at biological conditions, aminoglycosides bind to the negatively charged backbone of nucleic acids to disrupt protein synthesis, effectively inhibiting bacterial cell growth. APH(3') mediated phosphorylation of aminoglycosides effectively disrupts their mechanism of action, introducing a phosphate group that reduces their binding affinity due to steric hindrances and unfavorable electrostatic interactions. APH(3') is primarily found in certain species of gram-positive bacteria.
In enzymology, a nucleoside-triphosphate-aldose-1-phosphate nucleotidyltransferase is an enzyme that catalyzes the chemical reaction
CCA tRNA nucleotidyltransferase is an enzyme with systematic name CTP,CTP,ATP:tRNA cytidylyl,cytidylyl,adenylyltransferase. This enzyme catalyses the following chemical reaction

In molecular biology, the HEPN domain is a region of approximately 110 amino acids found in the C terminus of sacsin, a chaperonin implicated in an early-onset neurodegenerative disease in human, and in many bacterial and archaea proteins. There are three classes of proteins with HEPN domains:
2-deoxystreptamine N-acetyl-D-glucosaminyltransferase is an enzyme with systematic name UDP-N-acetyl-alpha-D-glucosamine:2-deoxystreptamine N-acetyl-D-glucosaminyltransferase. This enzyme catalyses the following chemical reaction
2-deoxystreptamine glucosyltransferase is an enzyme with systematic name UDP-alpha-D-glucose:2-deoxystreptamine 6-alpha-D-glucosyltransferase. This enzyme catalyses the following chemical reaction
The public domain consists of all the creative works to which no exclusive intellectual property rights apply. Those rights may have expired, been forfeited, expressly waived, or may be inapplicable.

Pfam is a database of protein families that includes their annotations and multiple sequence alignments generated using hidden Markov models. The most recent version, Pfam 32.0, was released in September 2018 and contains 17,929 families.
InterPro is a database of protein families, domains and functional sites in which identifiable features found in known proteins can be applied to new protein sequences in order to functionally characterise them.