
Nucleic acids are biopolymers, or large biomolecules, essential to all known forms of life. They are composed of nucleotides, which are the monomers made of three components: a 5-carbon sugar, a phosphate group and a nitrogenous base. The two main classes of nucleic acids are deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). If the sugar is ribose, the polymer is RNA; if the sugar is the ribose derivative deoxyribose, the polymer is DNA.

Nucleotides are organic molecules consisting of a nucleoside and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecules within all life-forms on Earth. Nucleotides are obtained in the diet and are also synthesized from common nutrients by the liver.
Deoxyribose, or more precisely 2-deoxyribose, is a monosaccharide with idealized formula H−(C=O)−(CH2)−(CHOH)3−H. Its name indicates that it is a deoxy sugar, meaning that it is derived from the sugar ribose by loss of an oxygen atom. Deoxyribose is most notable for its presence in DNA. Since the pentose sugars arabinose and ribose only differ by the stereochemistry at C2′, 2-deoxyribose and 2-deoxyarabinose are equivalent, although the latter term is rarely used because ribose, not arabinose, is the precursor to deoxyribose.

Adenosine monophosphate (AMP), also known as 5'-adenylic acid, is a nucleotide. AMP consists of a phosphate group, the sugar ribose, and the nucleobase adenine; it is an ester of phosphoric acid and the nucleoside adenosine. As a substituent it takes the form of the prefix adenylyl-.
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, research, and forensics. Commonly made in the laboratory by solid-phase chemical synthesis, these small bits of nucleic acids can be manufactured as single-stranded molecules with any user-specified sequence, and so are vital for artificial gene synthesis, polymerase chain reaction (PCR), DNA sequencing, molecular cloning and as molecular probes. In nature, oligonucleotides are usually found as small RNA molecules that function in the regulation of gene expression, or are degradation intermediates derived from the breakdown of larger nucleic acid molecules.

In biochemistry, a ribonucleotide is a nucleotide containing ribose as its pentose component. It is considered a molecular precursor of nucleic acids. Nucleotides are the basic building blocks of DNA and RNA. The monomer itself from ribonucleotides forms the basic building blocks for RNA. However, the reduction of ribonucleotide, by enzyme ribonucleotide reductase (RNR), forms deoxyribonucleotide, which is the essential building block for DNA. There are several differences between DNA deoxyribonucleotides and RNA ribonucleotides. Successive nucleotides are linked together via phosphodiester bonds by 3'-5'.

Leslie Eleazer Orgel FRS was a British chemist. He is known for his theories on the origin of life.
Aptamers are oligonucleotide or peptide molecules that bind to a specific target molecule. Aptamers are usually created by selecting them from a large random sequence pool, but natural aptamers also exist in riboswitches. Aptamers can be used for both basic research and clinical purposes as macromolecular drugs. Aptamers can be combined with ribozymes to self-cleave in the presence of their target molecule. These compound molecules have additional research, industrial and clinical applications.
Threose nucleic acid (TNA) is an artificial genetic polymer in which the natural five-carbon ribose sugar found in RNA has been replaced by an unnatural four-carbon threose sugar. Invented by Albert Eschenmoser as part of his quest to explore the chemical etiology of RNA, TNA has become an important synthetic genetic polymer (XNA) due to its ability to efficiently base pair with complementary sequences of DNA and RNA. However, unlike DNA and RNA, TNA is completely refractory to nuclease digestion, making it a promising nucleic acid analog for therapeutic and diagnostic applications.

Nucleic acid metabolism is the process by which nucleic acids are synthesized and degraded. Nucleic acids are the polymers of nucleotides. Nucleotide synthesis is an anabolic mechanism generally involving the chemical reaction of phosphate, pentose sugar, and a nitrogenous base. Destruction of nucleic acid is a catabolic reaction. Additionally, parts of the nucleotides or nucleobases can be salvaged to recreate new nucleotides. Both synthesis and degradation reactions require enzymes to facilitate the event. Defects or deficiencies in these enzymes can lead to a variety of diseases.

Systematic evolution of ligands by exponential enrichment (SELEX), also referred to as in vitro selection or in vitro evolution, is a combinatorial chemistry technique in molecular biology for producing oligonucleotides of either single-stranded DNA or RNA that specifically bind to a target ligand or ligands. These single-stranded DNA or RNA are commonly referred to as aptamers. Although SELEX has emerged as the most commonly used name for the procedure, some researchers have referred to it as SAAB and CASTing SELEX was first introduced in 1990. In 2015 a special issue was published in the Journal of Molecular Evolution in the honor of quarter century of the SELEX discovery.

Nucleic acid analogues are compounds which are analogous to naturally occurring RNA and DNA, used in medicine and in molecular biology research. Nucleic acids are chains of nucleotides, which are composed of three parts: a phosphate backbone, a pentose sugar, either ribose or deoxyribose, and one of four nucleobases. An analogue may have any of these altered. Typically the analogue nucleobases confer, among other things, different base pairing and base stacking properties. Examples include universal bases, which can pair with all four canonical bases, and phosphate-sugar backbone analogues such as PNA, which affect the properties of the chain . Nucleic acid analogues are also called Xeno Nucleic Acid and represent one of the main pillars of xenobiology, the design of new-to-nature forms of life based on alternative biochemistries.
A bridged nucleic acid (BNA) is a modified RNA nucleotide. They are sometimes also referred to as constrained or inaccessible RNA molecules. BNA monomers can contain a five-membered, six-membered or even a seven-membered bridged structure with a "fixed" C3'-endo sugar puckering. The bridge is synthetically incorporated at the 2', 4'-position of the ribose to afford a 2', 4'-BNA monomer. The monomers can be incorporated into oligonucleotide polymeric structures using standard phosphoamidite chemistry. BNAs are structurally rigid oligo-nucleotides with increased binding affinities and stability.

Xeno nucleic acids (XNA) are synthetic nucleic acid analogues that have a different sugar backbone than the natural nucleic acids DNA and RNA. As of 2011, at least six types of synthetic sugars have been shown to form nucleic acid backbones that can store and retrieve genetic information. Research is now being done to create synthetic polymerases to transform XNA. The study of its production and application has created a field known as xenobiology.

The need for fluorescently tracking RNA rose as its roles in complex cellular functions has grown to not only include mRNA, rRNA, and tRNA, but also RNAi, siRNA, snoRNA, and lncRNA, among others. Spinach is a synthetically derived RNA aptamer born out of the need for a way of studying the role of RNAs at the cellular level. This aptamer was created using Systematic Evolution for Ligands by EXponential enrichment, or SELEX, which is also known as in vitro evolution.

Optimer ligands are short synthetic oligonucleotide molecules composed of DNA or RNA that bind to a specific target molecule. They are engineered to bind their target molecules with affinity typically in the low nanomolar range. Optimers can be used as antibody mimetics in a range of applications, and have been optimized to increase their stability, reduce their molecular weight, and offer increased scalability and consistency in manufacture compared to standard aptamer molecules.
Mirror life is a hypothetical form of life with mirror-reflected molecular building blocks. The possibility of mirror life was first discussed by Louis Pasteur. Although this alternative life form has not been discovered in nature, efforts to build a mirror-image version of biology's molecular machinery are already underway.
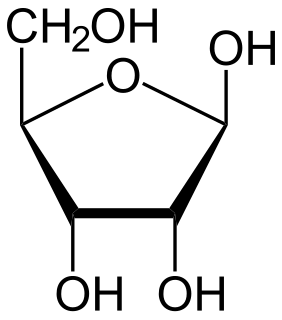
Ribose is a simple sugar and carbohydrate with molecular formula C5H10O5 and the linear-form composition H−(C=O)−(CHOH)4−H. The naturally-occurring form, d-ribose, is a component of the ribonucleotides from which RNA is built, and so this compound is necessary for coding, decoding, regulation and expression of genes. It has a structural analog, deoxyribose, which is a similarly essential component of DNA. l-ribose is an unnatural sugar that was first prepared by Emil Fischer and Oscar Piloty in 1891. It was not until 1909 that Phoebus Levene and Walter Jacobs recognised that d-ribose was a natural product, the enantiomer of Fischer and Piloty's product, and an essential component of nucleic acids. Fischer chose the name "ribose" as it is a partial rearrangement of the name of another sugar, arabinose, of which ribose is an epimer at the 2' carbon; both names also relate to gum arabic, from which arabinose was first isolated and from which they prepared l-ribose.
RNA therapeutics are a new class of medications based on ribonucleic acid (RNA). Research has been working on clinical use since the 1990s, with significant success in cancer therapy in the early 2010s. In 2020 and 2021, mRNA vaccines have been developed globally for use in combating the coronavirus disease. The Pfizer–BioNTech COVID-19 vaccine was the first mRNA vaccine approved by a medicines regulator, followed by the Moderna COVID-19 vaccine, and others.













