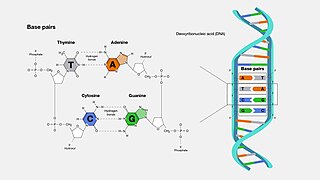
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA and RNA. Dictated by specific hydrogen bonding patterns, "Watson–Crick" base pairs allow the DNA helix to maintain a regular helical structure that is subtly dependent on its nucleotide sequence. The complementary nature of this based-paired structure provides a redundant copy of the genetic information encoded within each strand of DNA. The regular structure and data redundancy provided by the DNA double helix make DNA well suited to the storage of genetic information, while base-pairing between DNA and incoming nucleotides provides the mechanism through which DNA polymerase replicates DNA and RNA polymerase transcribes DNA into RNA. Many DNA-binding proteins can recognize specific base-pairing patterns that identify particular regulatory regions of genes.

Nucleic acids are large biomolecules that are crucial in all cells and viruses. They are composed of nucleotides, which are the monomer components: a 5-carbon sugar, a phosphate group and a nitrogenous base. The two main classes of nucleic acids are deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). If the sugar is ribose, the polymer is RNA; if the sugar is deoxyribose, a variant of ribose, the polymer is DNA.
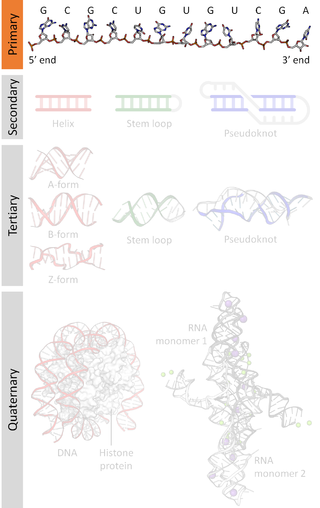
A nucleic acid sequence is a succession of bases within the nucleotides forming alleles within a DNA or RNA (GACU) molecule. This succession is denoted by a series of a set of five different letters that indicate the order of the nucleotides. By convention, sequences are usually presented from the 5' end to the 3' end. For DNA, with its double helix, there are two possible directions for the notated sequence; of these two, the sense strand is used. Because nucleic acids are normally linear (unbranched) polymers, specifying the sequence is equivalent to defining the covalent structure of the entire molecule. For this reason, the nucleic acid sequence is also termed the primary structure.

DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, guanine, cytosine, and thymine. The advent of rapid DNA sequencing methods has greatly accelerated biological and medical research and discovery.
Xenobiology (XB) is a subfield of synthetic biology, the study of synthesizing and manipulating biological devices and systems. The name "xenobiology" derives from the Greek word xenos, which means "stranger, alien". Xenobiology is a form of biology that is not (yet) familiar to science and is not found in nature. In practice, it describes novel biological systems and biochemistries that differ from the canonical DNA–RNA-20 amino acid system. For example, instead of DNA or RNA, XB explores nucleic acid analogues, termed xeno nucleic acid (XNA) as information carriers. It also focuses on an expanded genetic code and the incorporation of non-proteinogenic amino acids, or “xeno amino acids” into proteins.
In molecular biology, a hybridization probe (HP) is a fragment of DNA or RNA, usually 15–10000 nucleotides long, which can be radioactively or fluorescently labeled. HPs can be used to detect the presence of nucleotide sequences in analyzed RNA or DNA that are complementary to the sequence in the probe. The labeled probe is first denatured into single stranded DNA (ssDNA) and then hybridized to the target ssDNA or RNA immobilized on a membrane or in situ.

Hoechst stains are part of a family of blue fluorescent dyes used to stain DNA. These bis-benzimides were originally developed by Hoechst AG, which numbered all their compounds so that the dye Hoechst 33342 is the 33,342nd compound made by the company. There are three related Hoechst stains: Hoechst 33258, Hoechst 33342, and Hoechst 34580. The dyes Hoechst 33258 and Hoechst 33342 are the ones most commonly used and they have similar excitation–emission spectra.

Aminoallyl nucleotide is a nucleotide with a modified base containing an allylamine. They are used in post-labeling of nucleic acids by fluorescence detection in microarray. They are reactive with N-Hydroxysuccinimide ester group which helps attach a fluorescent dye to the primary amino group on the nucleotide. These nucleotides are known as 5-(3-aminoallyl)-nucleotides since the aminoallyl group is usually attached to carbon 5 of the pyrimidine ring of uracil or cytosine. The primary amine group in the aminoallyl moiety is aliphatic and thus more reactive compared to the amine groups that are directly attached to the rings (aromatic) of the bases. Common names of aminoallyl nucleosides are initially abbreviated with aa- or AA- to indicate aminoallyl. The 5-carbon sugar is indicated with or without the lowercase "d" indicating deoxyribose if included or ribose if not. Finally the nitrogenous base and number of phosphates are indicated.
Nucleic acid thermodynamics is the study of how temperature affects the nucleic acid structure of double-stranded DNA (dsDNA). The melting temperature (Tm) is defined as the temperature at which half of the DNA strands are in the random coil or single-stranded (ssDNA) state. Tm depends on the length of the DNA molecule and its specific nucleotide sequence. DNA, when in a state where its two strands are dissociated, is referred to as having been denatured by the high temperature.
Lipofectamine or Lipofectamine 2000 is a common transfection reagent, produced and sold by Invitrogen, used in molecular and cellular biology. It is used to increase the transfection efficiency of RNA or plasmid DNA into in vitro cell cultures by lipofection. Lipofectamine contains lipid subunits that can form liposomes in an aqueous environment, which entrap the transfection payload, e.g. DNA plasmids.
Applied Biosystems is one of various brands under the Life Technologies brand of Thermo Fisher Scientific corporation. The brand is focused on integrated systems for genetic analysis, which include computerized machines and the consumables used within them.

Non-histone chromosomal protein HMG-14 is a protein that in humans is encoded by the HMGN1 gene.

Transcription factor SOX-6 is a protein that in humans is encoded by the SOX6 gene.
The nucleic acid notation currently in use was first formalized by the International Union of Pure and Applied Chemistry (IUPAC) in 1970. This universally accepted notation uses the Roman characters G, C, A, and T, to represent the four nucleotides commonly found in deoxyribonucleic acids (DNA).
The Qubit fluorometer is a laboratory instrument developed and distributed by Invitrogen, which is now a part of Thermo Fisher. It is used for the quantification of DNA, RNA, and protein.
TRANSFAC is a manually curated database of eukaryotic transcription factors, their genomic binding sites and DNA binding profiles. The contents of the database can be used to predict potential transcription factor binding sites.

The European Nucleotide Archive (ENA) is a repository providing free and unrestricted access to annotated DNA and RNA sequences. It also stores complementary information such as experimental procedures, details of sequence assembly and other metadata related to sequencing projects. The archive is composed of three main databases: the Sequence Read Archive, the Trace Archive and the EMBL Nucleotide Sequence Database. The ENA is produced and maintained by the European Bioinformatics Institute and is a member of the International Nucleotide Sequence Database Collaboration (INSDC) along with the DNA Data Bank of Japan and GenBank.
Tom Brown FRSC FRSE is a British chemist, biotechnologist, and entrepreneur. He is the Professor of Nucleic acid chemistry at the Department of Chemistry and Department of Oncology at the University of Oxford. Currently, he is serving as the President of the Chemical Biology Interface Division of the Royal Society of Chemistry. He is best known for his contribution in the field of DNA Repair, DNA Click chemistry, and in the application of Molecular genetics in forensics and diagnostics.








