
The RNA world is a hypothetical stage in the evolutionary history of life on Earth, in which self-replicating RNA molecules proliferated before the evolution of DNA and proteins. The term also refers to the hypothesis that posits the existence of this stage.
Ribozymes are RNA molecules that have the ability to catalyze specific biochemical reactions, including RNA splicing in gene expression, similar to the action of protein enzymes. The 1982 discovery of ribozymes demonstrated that RNA can be both genetic material and a biological catalyst, and contributed to the RNA world hypothesis, which suggests that RNA may have been important in the evolution of prebiotic self-replicating systems.
Gene silencing is the regulation of gene expression in a cell to prevent the expression of a certain gene. Gene silencing can occur during either transcription or translation and is often used in research. In particular, methods used to silence genes are being increasingly used to produce therapeutics to combat cancer and other diseases, such as infectious diseases and neurodegenerative disorders.
Virusoids are circular single-stranded RNA(s) dependent on viruses for replication and encapsidation. The genome of virusoids consist of several hundred (200–400) nucleotides and does not code for any proteins.
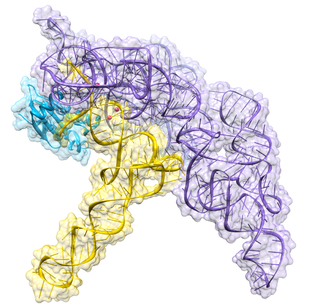
Ribonuclease P is a type of ribonuclease which cleaves RNA. RNase P is unique from other RNases in that it is a ribozyme – a ribonucleic acid that acts as a catalyst in the same way that a protein-based enzyme would. Its function is to cleave off an extra, or precursor, sequence of RNA on tRNA molecules. Further, RNase P is one of two known multiple turnover ribozymes in nature, the discovery of which earned Sidney Altman and Thomas Cech the Nobel Prize in Chemistry in 1989: in the 1970s, Altman discovered the existence of precursor tRNA with flanking sequences and was the first to characterize RNase P and its activity in processing of the 5' leader sequence of precursor tRNA. Recent findings also reveal that RNase P has a new function. It has been shown that human nuclear RNase P is required for the normal and efficient transcription of various small noncoding RNAs, such as tRNA, 5S rRNA, SRP RNA and U6 snRNA genes, which are transcribed by RNA polymerase III, one of three major nuclear RNA polymerases in human cells.
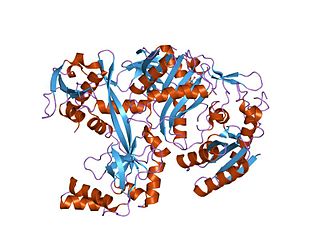
Piwi genes were identified as regulatory proteins responsible for stem cell and germ cell differentiation. Piwi is an abbreviation of P-elementInduced WImpy testis in Drosophila. Piwi proteins are highly conserved RNA-binding proteins and are present in both plants and animals. Piwi proteins belong to the Argonaute/Piwi family and have been classified as nuclear proteins. Studies on Drosophila have also indicated that Piwi proteins have no slicer activity conferred by the presence of the Piwi domain. In addition, Piwi associates with heterochromatin protein 1, an epigenetic modifier, and piRNA-complementary sequences. These are indications of the role Piwi plays in epigenetic regulation. Piwi proteins are also thought to control the biogenesis of piRNA as many Piwi-like proteins contain slicer activity which would allow Piwi proteins to process precursor piRNA into mature piRNA.
RNA-induced transcriptional silencing (RITS) is a form of RNA interference by which short RNA molecules – such as small interfering RNA (siRNA) – trigger the downregulation of transcription of a particular gene or genomic region. This is usually accomplished by posttranslational modification of histone tails which target the genomic region for heterochromatin formation. The protein complex that binds to siRNAs and interacts with the methylated lysine 9 residue of histones H3 (H3K9me2) is the RITS complex.

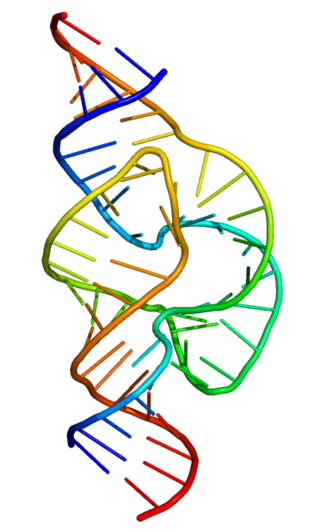
The hammerhead ribozyme is an RNA motif that catalyzes reversible cleavage and ligation reactions at a specific site within an RNA molecule. It is one of several catalytic RNAs (ribozymes) known to occur in nature. It serves as a model system for research on the structure and properties of RNA, and is used for targeted RNA cleavage experiments, some with proposed therapeutic applications. Named for the resemblance of early secondary structure diagrams to a hammerhead shark, hammerhead ribozymes were originally discovered in two classes of plant virus-like RNAs: satellite RNAs and viroids. They are also known in some classes of retrotransposons, including the retrozymes. The hammerhead ribozyme motif has been ubiquitously reported in lineages across the tree of life.

The hairpin ribozyme is a small section of RNA that can act as a ribozyme. Like the hammerhead ribozyme it is found in RNA satellites of plant viruses. It was first identified in the minus strand of the tobacco ringspot virus (TRSV) satellite RNA where it catalyzes self-cleavage and joining (ligation) reactions to process the products of rolling circle virus replication into linear and circular satellite RNA molecules. The hairpin ribozyme is similar to the hammerhead ribozyme in that it does not require a metal ion for the reaction.

The glucosamine-6-phosphate riboswitch ribozyme is an RNA structure that resides in the 5' untranslated region (UTR) of the mRNA transcript of the glmS gene. This RNA regulates the glmS gene by responding to concentrations of a specific metabolite, glucosamine-6-phosphate (GlcN6P), in addition to catalyzing a self-cleaving chemical reaction upon activation. This cleavage leads to the degradation of the mRNA that contains the ribozyme, and lowers production of GlcN6P. The glmS gene encodes for an enzyme glutamine-fructose-6-phosphate amidotransferase, which catalyzes the formation of GlcN6P, a compound essential for cell wall biosynthesis, from fructose-6-phosphate and glutamine. Thus, when GlcN6P levels are high, the glmS ribozyme is activated and the mRNA transcript is degraded but in the absence of GlcN6P the gene continues to be translated into glutamine-fructose-6-phosphate amidotransferase and GlcN6P is produced. GlcN6P is a cofactor for this cleavage reaction, as it directly participates as an acid-base catalyst. This RNA is the first riboswitch also found to be a self-cleaving ribozyme and, like many others, was discovered using a bioinformatics approach.
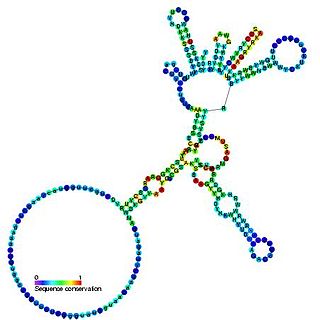
Group I introns are large self-splicing ribozymes. They catalyze their own excision from mRNA, tRNA and rRNA precursors in a wide range of organisms. The core secondary structure consists of nine paired regions (P1-P9). These fold to essentially two domains – the P4-P6 domain and the P3-P9 domain. The secondary structure mark-up for this family represents only this conserved core. Group I introns often have long open reading frames inserted in loop regions.

The hepatitis delta virus (HDV) ribozyme is a non-coding RNA found in the hepatitis delta virus that is necessary for viral replication and is the only known human virus that utilizes ribozyme activity to infect its host. The ribozyme acts to process the RNA transcripts to unit lengths in a self-cleavage reaction during replication of the hepatitis delta virus, which is thought to propagate by a double rolling circle mechanism. The ribozyme is active in vivo in the absence of any protein factors and was the fastest known naturally occurring self-cleaving RNA at the time of its discovery.
CPEB, or cytoplasmic polyadenylation element binding protein, is a highly conserved RNA-binding protein that promotes the elongation of the polyadenine tail of messenger RNA. CPEB most commonly activates the target RNA for translation, but can also act as a repressor, dependent on its phosphorylation state. In animals, CPEB is expressed in several alternative splicing isoforms that are specific to particular tissues and functions, including the self-cleaving Mammalian CPEB3 ribozyme. CPEB was first identified in Xenopus oocytes and associated with meiosis; a role has also been identified in the spermatogenesis of Caenorhabditis elegans.

Origin recognition complex subunit 6 is a protein that in humans is encoded by the ORC6 (ORC6L) gene.
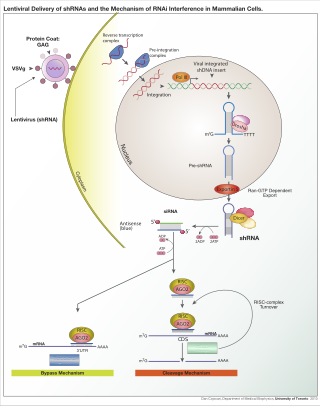
RNA interference (RNAi) is a biological process in which RNA molecules are involved in sequence-specific suppression of gene expression by double-stranded RNA, through translational or transcriptional repression. Historically, RNAi was known by other names, including co-suppression, post-transcriptional gene silencing (PTGS), and quelling. The detailed study of each of these seemingly different processes elucidated that the identity of these phenomena were all actually RNAi. Andrew Fire and Craig C. Mello shared the 2006 Nobel Prize in Physiology or Medicine for their work on RNAi in the nematode worm Caenorhabditis elegans, which they published in 1998. Since the discovery of RNAi and its regulatory potentials, it has become evident that RNAi has immense potential in suppression of desired genes. RNAi is now known as precise, efficient, stable and better than antisense therapy for gene suppression. Antisense RNA produced intracellularly by an expression vector may be developed and find utility as novel therapeutic agents.
David P. Bartel is an American molecular biologist best known for his work on microRNAs. Bartel is a Professor of Biology at the Massachusetts Institute of Technology, Member of the Whitehead Institute, and investigator of the Howard Hughes Medical Institute (HHMI).

The twister ribozyme is a catalytic RNA structure capable of self-cleavage. The nucleolytic activity of this ribozyme has been demonstrated both in vivo and in vitro and has one of the fastest catalytic rates of naturally occurring ribozymes with similar function. The twister ribozyme is considered to be a member of the small self-cleaving ribozyme family which includes the hammerhead, hairpin, hepatitis delta virus (HDV), Varkud satellite (VS), and glmS ribozymes.

The microprocessor complex is a protein complex involved in the early stages of processing microRNA (miRNA) and RNA interference (RNAi) in animal cells. The complex is minimally composed of the ribonuclease enzyme Drosha and the dimeric RNA-binding protein DGCR8, and cleaves primary miRNA substrates to pre-miRNA in the cell nucleus. Microprocessor is also the smaller of the two multi-protein complexes that contain human Drosha.

RNAs Associated with Genes Associated with Twister and Hammerhead ribozymes (RAGATH) refers to a bioinformatics strategy that was devised to find self-cleaving ribozymes in bacteria. It also refers to candidate RNAs, or RAGATH RNA motifs, discovered using this strategy.

Hovlinc RNA is a self-cleaving ribozyme of about 168 nucleotides found in a very long noncoding RNA (vlincRNA) in humans, chimpanzees, and gorillas. The word "hovlinc" comes from "hominin vlincRNA-located" RNA. Hovlinc is only a fourth known case of a ribozyme in human. Self-cleavage activity of Hovlinc has been shown in human, chimpanzees and bonobos, but is absent in gorillas, raising questions about Hovlinc's biological function and evolution.















