
In biology, epigenetics is the study of heritable traits, or a stable change of cell function, that happen without changes to the DNA sequence. The Greek prefix epi- in epigenetics implies features that are "on top of" or "in addition to" the traditional genetic mechanism of inheritance. Epigenetics usually involves a change that is not erased by cell division, and affects the regulation of gene expression. Such effects on cellular and physiological phenotypic traits may result from environmental factors, or be part of normal development. Epigenetic factors can also lead to cancer.

Regulation of gene expression, or gene regulation, includes a wide range of mechanisms that are used by cells to increase or decrease the production of specific gene products. Sophisticated programs of gene expression are widely observed in biology, for example to trigger developmental pathways, respond to environmental stimuli, or adapt to new food sources. Virtually any step of gene expression can be modulated, from transcriptional initiation, to RNA processing, and to the post-translational modification of a protein. Often, one gene regulator controls another, and so on, in a gene regulatory network.

In biology, the epigenome of an organism is the collection of chemical changes to its DNA and histone proteins that affects when, where, and how the DNA is expressed; these changes can be passed down to an organism's offspring via transgenerational epigenetic inheritance. Changes to the epigenome can result in changes to the structure of chromatin and changes to the function of the genome. The human epigenome, including DNA methylation and histone modification, is maintained through cell division. The epigenome is essential for normal development and cellular differentiation, enabling cells with the same genetic code to perform different functions. The human epigenome is dynamic and can be influenced by environmental factors such as diet, stress, and toxins.
Histone methylation is a process by which methyl groups are transferred to amino acids of histone proteins that make up nucleosomes, which the DNA double helix wraps around to form chromosomes. Methylation of histones can either increase or decrease transcription of genes, depending on which amino acids in the histones are methylated, and how many methyl groups are attached. Methylation events that weaken chemical attractions between histone tails and DNA increase transcription because they enable the DNA to uncoil from nucleosomes so that transcription factor proteins and RNA polymerase can access the DNA. This process is critical for the regulation of gene expression that allows different cells to express different genes.

Methyltransferases are a large group of enzymes that all methylate their substrates but can be split into several subclasses based on their structural features. The most common class of methyltransferases is class I, all of which contain a Rossmann fold for binding S-Adenosyl methionine (SAM). Class II methyltransferases contain a SET domain, which are exemplified by SET domain histone methyltransferases, and class III methyltransferases, which are membrane associated. Methyltransferases can also be grouped as different types utilizing different substrates in methyl transfer reactions. These types include protein methyltransferases, DNA/RNA methyltransferases, natural product methyltransferases, and non-SAM dependent methyltransferases. SAM is the classical methyl donor for methyltransferases, however, examples of other methyl donors are seen in nature. The general mechanism for methyl transfer is a SN2-like nucleophilic attack where the methionine sulfur serves as the leaving group and the methyl group attached to it acts as the electrophile that transfers the methyl group to the enzyme substrate. SAM is converted to S-Adenosyl homocysteine (SAH) during this process. The breaking of the SAM-methyl bond and the formation of the substrate-methyl bond happen nearly simultaneously. These enzymatic reactions are found in many pathways and are implicated in genetic diseases, cancer, and metabolic diseases. Another type of methyl transfer is the radical S-Adenosyl methionine (SAM) which is the methylation of unactivated carbon atoms in primary metabolites, proteins, lipids, and RNA.

Histone-modifying enzymes are enzymes involved in the modification of histone substrates after protein translation and affect cellular processes including gene expression. To safely store the eukaryotic genome, DNA is wrapped around four core histone proteins, which then join to form nucleosomes. These nucleosomes further fold together into highly condensed chromatin, which renders the organism's genetic material far less accessible to the factors required for gene transcription, DNA replication, recombination and repair. Subsequently, eukaryotic organisms have developed intricate mechanisms to overcome this repressive barrier imposed by the chromatin through histone modification, a type of post-translational modification which typically involves covalently attaching certain groups to histone residues. Once added to the histone, these groups elicit either a loose and open histone conformation, euchromatin, or a tight and closed histone conformation, heterochromatin. Euchromatin marks active transcription and gene expression, as the light packing of histones in this way allows entry for proteins involved in the transcription process. As such, the tightly packed heterochromatin marks the absence of current gene expression.
Chromatin remodeling is the dynamic modification of chromatin architecture to allow access of condensed genomic DNA to the regulatory transcription machinery proteins, and thereby control gene expression. Such remodeling is principally carried out by 1) covalent histone modifications by specific enzymes, e.g., histone acetyltransferases (HATs), deacetylases, methyltransferases, and kinases, and 2) ATP-dependent chromatin remodeling complexes which either move, eject or restructure nucleosomes. Besides actively regulating gene expression, dynamic remodeling of chromatin imparts an epigenetic regulatory role in several key biological processes, egg cells DNA replication and repair; apoptosis; chromosome segregation as well as development and pluripotency. Aberrations in chromatin remodeling proteins are found to be associated with human diseases, including cancer. Targeting chromatin remodeling pathways is currently evolving as a major therapeutic strategy in the treatment of several cancers.
Computational epigenetics uses statistical methods and mathematical modelling in epigenetic research. Due to the recent explosion of epigenome datasets, computational methods play an increasing role in all areas of epigenetic research.

Siddhartha Mukherjee is an Indian-American physician, biologist, and author. He is best known for his 2010 book, The Emperor of All Maladies: A Biography of Cancer, that won notable literary prizes including the 2011 Pulitzer Prize for General Non-Fiction, and Guardian First Book Award, among others. The book was listed in the "All-Time 100 Nonfiction Books" by Time magazine in 2011. His 2016 book The Gene: An Intimate History made it to #1 on The New York Times Best Seller list, and was among The New York Times 100 best books of 2016, and a finalist for the Wellcome Trust Prize and the Royal Society Prize for Science Books.

Cancer epigenetics is the study of epigenetic modifications to the DNA of cancer cells that do not involve a change in the nucleotide sequence, but instead involve a change in the way the genetic code is expressed. Epigenetic mechanisms are necessary to maintain normal sequences of tissue specific gene expression and are crucial for normal development. They may be just as important, if not even more important, than genetic mutations in a cell's transformation to cancer. The disturbance of epigenetic processes in cancers, can lead to a loss of expression of genes that occurs about 10 times more frequently by transcription silencing than by mutations. As Vogelstein et al. points out, in a colorectal cancer there are usually about 3 to 6 driver mutations and 33 to 66 hitchhiker or passenger mutations. However, in colon tumors compared to adjacent normal-appearing colonic mucosa, there are about 600 to 800 heavily methylated CpG islands in the promoters of genes in the tumors while these CpG islands are not methylated in the adjacent mucosa. Manipulation of epigenetic alterations holds great promise for cancer prevention, detection, and therapy. In different types of cancer, a variety of epigenetic mechanisms can be perturbed, such as the silencing of tumor suppressor genes and activation of oncogenes by altered CpG island methylation patterns, histone modifications, and dysregulation of DNA binding proteins. There are several medications which have epigenetic impact, that are now used in a number of these diseases.
The epigenetics of schizophrenia is the study of how inherited epigenetic changes are regulated and modified by the environment and external factors and how these changes influence the onset and development of, and vulnerability to, schizophrenia. Epigenetics concerns the heritability of those changes, too. Schizophrenia is a debilitating and often misunderstood disorder that affects up to 1% of the world's population. Although schizophrenia is a heavily studied disorder, it has remained largely impervious to scientific understanding; epigenetics offers a new avenue for research, understanding, and treatment.
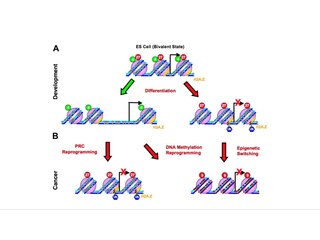
Epigenetic therapy refers to the use of drugs or other interventions to modify gene expression patterns, potentially treating diseases by targeting epigenetic mechanisms such as DNA methylation and histone modifications.

Nessa Carey is a British biologist working in the field of molecular biology and biotechnology. She is International Director of the technology transfer organization PraxisUnico and a visiting professor at Imperial College London.
Epigenetics of human development is the study of how epigenetics effects human development.
Neuroepigenetics is the study of how epigenetic changes to genes affect the nervous system. These changes may effect underlying conditions such as addiction, cognition, and neurological development.
H3K27me3 is an epigenetic modification to the DNA packaging protein histone H3. It is a mark that indicates the tri-methylation of lysine 27 on histone H3 protein.

An epigenome-wide association study (EWAS) is an examination of a genome-wide set of quantifiable epigenetic marks, such as DNA methylation, in different individuals to derive associations between epigenetic variation and a particular identifiable phenotype/trait. When patterns change such as DNA methylation at specific loci, discriminating the phenotypically affected cases from control individuals, this is considered an indication that epigenetic perturbation has taken place that is associated, causally or consequentially, with the phenotype.
CpG island hypermethylation is a phenomenon that is important for the regulation of gene expression in cancer cells, as an epigenetic control aberration responsible for gene inactivation. Hypermethylation of CpG islands has been described in almost every type of tumor.
Pharmacoepigenetics is an emerging field that studies the underlying epigenetic marking patterns that lead to variation in an individual's response to medical treatment.
Epigenetics in forensic science is the application of epigenetics to solving crimes.











