Related Research Articles

Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. These products are often proteins, but in non-protein-coding genes such as transfer RNA (tRNA) and small nuclear RNA (snRNA), the product is a functional non-coding RNA. Gene expression is summarized in the central dogma of molecular biology first formulated by Francis Crick in 1958, further developed in his 1970 article, and expanded by the subsequent discoveries of reverse transcription and RNA replication.

A non-coding RNA (ncRNA) is an RNA molecule that is not translated into a protein. The DNA sequence from which a functional non-coding RNA is transcribed is often called an RNA gene. Abundant and functionally important types of non-coding RNAs include transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs), as well as small RNAs such as microRNAs, siRNAs, piRNAs, snoRNAs, snRNAs, exRNAs, scaRNAs and the long ncRNAs such as Xist and HOTAIR.

Alternative splicing, or alternative RNA splicing, or differential splicing, is an alternative splicing process during gene expression that allows a single gene to code for multiple proteins. In this process, particular exons of a gene may be included within or excluded from the final, processed messenger RNA (mRNA) produced from that gene. This means the exons are joined in different combinations, leading to different (alternative) mRNA strands. Consequently, the proteins translated from alternatively spliced mRNAs will contain differences in their amino acid sequence and, often, in their biological functions. Notably, alternative splicing allows the human genome to direct the synthesis of many more proteins than would be expected from its 20,000 protein-coding genes.
A regulatory sequence is a segment of a nucleic acid molecule which is capable of increasing or decreasing the expression of specific genes within an organism. Regulation of gene expression is an essential feature of all living organisms and viruses.

A generegulatory network (GRN) is a collection of molecular regulators that interact with each other and with other substances in the cell to govern the gene expression levels of mRNA and proteins which, in turn, determine the function of the cell. GRN also play a central role in morphogenesis, the creation of body structures, which in turn is central to evolutionary developmental biology (evo-devo).

Regulation of gene expression, or gene regulation, includes a wide range of mechanisms that are used by cells to increase or decrease the production of specific gene products. Sophisticated programs of gene expression are widely observed in biology, for example to trigger developmental pathways, respond to environmental stimuli, or adapt to new food sources. Virtually any step of gene expression can be modulated, from transcriptional initiation, to RNA processing, and to the post-translational modification of a protein. Often, one gene regulator controls another, and so on, in a gene regulatory network.
RNA-binding proteins are proteins that bind to the double or single stranded RNA in cells and participate in forming ribonucleoprotein complexes. RBPs contain various structural motifs, such as RNA recognition motif (RRM), dsRNA binding domain, zinc finger and others. They are cytoplasmic and nuclear proteins. However, since most mature RNA is exported from the nucleus relatively quickly, most RBPs in the nucleus exist as complexes of protein and pre-mRNA called heterogeneous ribonucleoprotein particles (hnRNPs). RBPs have crucial roles in various cellular processes such as: cellular function, transport and localization. They especially play a major role in post-transcriptional control of RNAs, such as: splicing, polyadenylation, mRNA stabilization, mRNA localization and translation. Eukaryotic cells express diverse RBPs with unique RNA-binding activity and protein–protein interaction. According to the Eukaryotic RBP Database (EuRBPDB), there are 2961 genes encoding RBPs in humans. During evolution, the diversity of RBPs greatly increased with the increase in the number of introns. Diversity enabled eukaryotic cells to utilize RNA exons in various arrangements, giving rise to a unique RNP (ribonucleoprotein) for each RNA. Although RBPs have a crucial role in post-transcriptional regulation in gene expression, relatively few RBPs have been studied systematically.
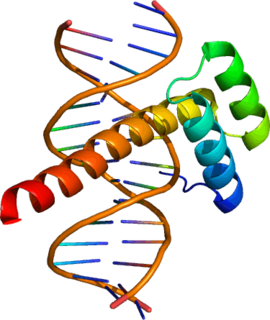
Ultrabithorax (Ubx) is a homeobox gene found in insects, and is used in the regulation of patterning in morphogenesis. There are many possible products of this gene, which function as transcription factors. Ubx is used in the specification of serially homologous structures, and is used at many levels of developmental hierarchies. In Drosophila melanogaster it is expressed in the third thoracic (T3) and first abdominal (A1) segments and represses wing formation. The Ubx gene regulates the decisions regarding the number of wings and legs the adult flies will have. The developmental role of the Ubx gene is determined by the splicing of its product, which takes place after translation of the gene. The specific splice factors of a particular cell allow the specific regulation of the developmental fate of that cell, by making different splice variants of transcription factors. In D. melanogaster, at least six different isoforms of Ubx exist.

Twist-related protein 1 (TWIST1) also known as class A basic helix–loop–helix protein 38 (bHLHa38) is a basic helix-loop-helix transcription factor that in humans is encoded by the TWIST1 gene.

Microphthalmia-associated transcription factor also known as class E basic helix-loop-helix protein 32 or bHLHe32 is a protein that in humans is encoded by the MITF gene.

High-mobility group AT-hook 2, also known as HMGA2, is a protein that, in humans, is encoded by the HMGA2 gene.

Metastasis-associated protein MTA1 is a protein that in humans is encoded by the MTA1 gene. MTA1 is the founding member of the MTA family of genes. MTA1 is primarily localized in the nucleus but also found to be distributed in the extra-nuclear compartments. MTA1 is a component of several chromatin remodeling complexes including the nucleosome remodeling and deacetylation complex (NuRD). MTA1 regulates gene expression by functioning as a coregulator to integrate DNA-interacting factors to gene activity. MTA1 participates in physiological functions in the normal and cancer cells. MTA1 is one of the most upregulated proteins in human cancer and associates with cancer progression, aggressive phenotypes, and poor prognosis of cancer patients.

Paired box gene 8, also known as PAX8, is a protein which in humans is encoded by the PAX8 gene.

Paired-like homeodomain transcription factor 2 also known as pituitary homeobox 2 is a protein that in humans is encoded by the PITX2 gene.

Forkhead box C1, also known as FOXC1, is a protein which in humans is encoded by the FOXC1 gene.

Forkhead box protein O4 is a protein that in humans is encoded by the FOXO4 gene.

T-box transcription factor TBX3 is a protein that in humans is encoded by the TBX3 gene.

Metastasis-associated protein MTA3 is a protein that in humans is encoded by the MTA3 gene. MTA3 protein localizes in the nucleus as well as in other cellular compartments MTA3 is a component of the nucleosome remodeling and deacetylate (NuRD) complex and participates in gene expression. The expression pattern of MTA3 is opposite to that of MTA1 and MTA2 during mammary gland tumorigenesis. However, MTA3 is also overexpressed in a variety of human cancers.

MALAT 1 also known as NEAT2 is a large, infrequently spliced non-coding RNA, which is highly conserved amongst mammals and highly expressed in the nucleus. MALAT1 was identified in multiple types of physiological processes, such as alternative splicing, nuclear organization, epigenetic modulating of gene expression, and a number of evidences indicate that MALAT1 also closely relate to various pathological processes, ranging from diabetes complications to cancers. It regulates the expression of metastasis-associated genes. It also positively regulates cell motility via the transcriptional and/or post-transcriptional regulation of motility-related genes. MALAT1 may play a role in temperature-dependent sex determination in the Red-eared slider turtle.

Forkhead box protein J1 is a protein that in humans is encoded by the FOXJ1 gene. It is a member of the Forkhead/winged helix (FOX) family of transcription factors that is involved in ciliogenesis. FOXJ1 is expressed in ciliated cells of the lung, choroid plexus, reproductive tract, embryonic kidney and pre-somite embryo stage.
References
- 1 2 Mattick, JS; Taft, RJ; Faulkner, GJ (January 2010). "A global view of genomic information--moving beyond the gene and the master regulator". Trends in Genetics. 26 (1): 21–8. doi:10.1016/j.tig.2009.11.002. PMID 19944475.
- ↑ Yang, J; Mani, SA; Donaher, JL; Ramaswamy, S; Itzykson, RA; Come, C; Savagner, P; Gitelman, I; Richardson, A; Weinberg, RA (25 June 2004). "Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis". Cell. 117 (7): 927–39. doi: 10.1016/j.cell.2004.06.006 . PMID 15210113. S2CID 16181905.
- ↑ Long, JC; Caceres, JF (1 January 2009). "The SR protein family of splicing factors: master regulators of gene expression". The Biochemical Journal. 417 (1): 15–27. doi:10.1042/BJ20081501. PMID 19061484.
- ↑ Brennecke, J; Aravin, AA; Stark, A; Dus, M; Kellis, M; Sachidanandam, R; Hannon, GJ (23 March 2007). "Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila" (PDF). Cell. 128 (6): 1089–103. doi: 10.1016/j.cell.2007.01.043 . PMID 17346786. S2CID 2246942.
- ↑ Oestreich, KJ; Weinmann, AS (November 2012). "Master regulators or lineage-specifying? Changing views on CD4+ T cell transcription factors". Nature Reviews. Immunology. 12 (11): 799–804. doi:10.1038/nri3321. PMC 3584691 . PMID 23059426.