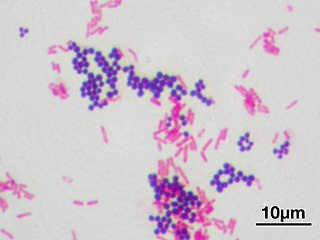
In microbiology and bacteriology, Gram stain, is a method of staining used to classify bacterial species into two large groups: gram-positive bacteria and gram-negative bacteria. The name comes from the Danish bacteriologist Hans Christian Gram, who developed the technique in 1884.
In cell biology, an organelle is a specialized subunit, usually within a cell, that has a specific function. The name organelle comes from the idea that these structures are parts of cells, as organs are to the body, hence organelle, the suffix -elle being a diminutive. Organelles are either separately enclosed within their own lipid bilayers or are spatially distinct functional units without a surrounding lipid bilayer. Although most organelles are functional units within cells, some function units that extend outside of cells are often termed organelles, such as cilia, the flagellum and archaellum, and the trichocyst.

The periplasm is a concentrated gel-like matrix in the space between the inner cytoplasmic membrane and the bacterial outer membrane called the periplasmic space in gram-negative bacteria. Using cryo-electron microscopy it has been found that a much smaller periplasmic space is also present in gram-positive bacteria, between cell wall and the plasma membrane.
An S-layer is a part of the cell envelope found in almost all archaea, as well as in many types of bacteria. The S-layers of both archaea and bacteria consists of a monomolecular layer composed of only one identical proteins or glycoproteins. This structure is built via self-assembly and encloses the whole cell surface. Thus, the S-layer protein can represent up to 15% of the whole protein content of a cell. S-layer proteins are poorly conserved or not conserved at all, and can differ markedly even between related species. Depending on species, the S-layers have a thickness between 5 and 25 nm and possess identical pores with 2–8 nm in diameter.
Azotobacter vinelandii is Gram-negative diazotroph that can fix nitrogen while grown aerobically. These bacteria are easily cultured and grown.

The bacterial outer membrane is found in gram-negative bacteria. Its composition is distinct from that of the inner cytoplasmic cell membrane - among other things, the outer leaflet of the outer membrane of many gram-negative bacteria includes a complex lipopolysaccharide whose lipid portion acts as an endotoxin - and in some bacteria such as E. coli it is linked to the cell's peptidoglycan by Braun's lipoprotein.

Electron cryotomography (CryoET) is an imaging technique used to produce high-resolution (~1–4 nm) three-dimensional views of samples, often biological macromolecules and cells. CryoET is a specialized application of transmission electron cryomicroscopy (CryoTEM) in which samples are imaged as they are tilted, resulting in a series of 2D images that can be combined to produce a 3D reconstruction, similar to a CT scan of the human body. In contrast to other electron tomography techniques, samples are imaged under cryogenic conditions. For cellular material, the structure is immobilized in non-crystalline, vitreous ice, allowing them to be imaged without dehydration or chemical fixation, which would otherwise disrupt or distort biological structures.
Members of the genus Selenomonas are referred to trivially as selenomonads. The genus Selenomonas constitutes a group of motile crescent-shaped bacteria and includes species living in the gastrointestinal tracts of animals, in particular the ruminants. A number of smaller forms discovered with the light microscope are now in culture but many, especially the large selenomonads are not, owing to their fastidious and incompletely known growth requirements.
Penicillin-binding proteins (PBPs) are a group of proteins that are characterized by their affinity for and binding of penicillin. They are a normal constituent of many bacteria; the name just reflects the way by which the protein was discovered. All β-lactam antibiotics bind to PBPs, which are essential for bacterial cell wall synthesis. PBPs are members of a subgroup of enzymes called transpeptidases. Specifically, PBPs are DD-transpeptidases.
The bacterium, despite its simplicity, contains a well-developed cell structure which is responsible for some of its unique biological structures and pathogenicity. Many structural features are unique to bacteria and are not found among archaea or eukaryotes. Because of the simplicity of bacteria relative to larger organisms and the ease with which they can be manipulated experimentally, the cell structure of bacteria has been well studied, revealing many biochemical principles that have been subsequently applied to other organisms.

Rhodospirillum rubrum is a Gram-negative, pink-coloured bacterium, with a size of 800 to 1000 nanometers. It is a facultative anaerobe, thus capable of using oxygen for aerobic respiration under aerobic conditions, or an alternative terminal electron acceptor for anaerobic respiration under anaerobic conditions. Alternative terminal electron acceptors for R. rubrum include dimethyl sulfoxide or trimethylamine oxide.
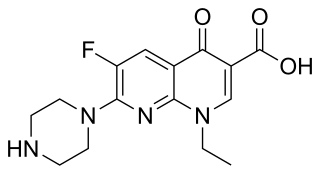
Enoxacin is an oral broad-spectrum fluoroquinolone antibacterial agent used in the treatment of urinary tract infections and gonorrhea. Insomnia is a common adverse effect. It is no longer available in the United States.
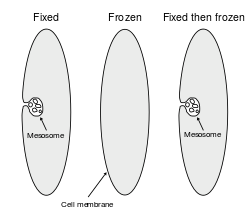
In the fields of histology, pathology, and cell biology, fixation is the preservation of biological tissues from decay due to autolysis or putrefaction. It terminates any ongoing biochemical reactions and may also increase the treated tissues' mechanical strength or stability. Tissue fixation is a critical step in the preparation of histological sections, its broad objective being to preserve cells and tissue components and to do this in such a way as to allow for the preparation of thin, stained sections. This allows the investigation of the tissues' structure, which is determined by the shapes and sizes of such macromolecules as proteins and nucleic acids.

Bacteria are ubiquitous, mostly free-living organisms often consisting of one biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were among the first life forms to appear on Earth, and are present in most of its habitats. Bacteria inhabit soil, water, acidic hot springs, radioactive waste, and the deep biosphere of Earth's crust. Bacteria play a vital role in many stages of the nutrient cycle by recycling nutrients and the fixation of nitrogen from the atmosphere. The nutrient cycle includes the decomposition of dead bodies; bacteria are responsible for the putrefaction stage in this process. In the biological communities surrounding hydrothermal vents and cold seeps, extremophile bacteria provide the nutrients needed to sustain life by converting dissolved compounds, such as hydrogen sulphide and methane, to energy. Bacteria also live in symbiotic and parasitic relationships with plants and animals. Most bacteria have not been characterised and there are many species that cannot be grown in the laboratory. The study of bacteria is known as bacteriology, a branch of microbiology.
Autoinducers are signaling molecules that are produced in response to changes in cell-population density. As the density of quorum sensing bacterial cells increases so does the concentration of the autoinducer. Detection of signal molecules by bacteria acts as stimulation which leads to altered gene expression once the minimal threshold is reached. Quorum sensing is a phenomenon that allows both Gram-negative and Gram-positive bacteria to sense one another and to regulate a wide variety of physiological activities. Such activities include symbiosis, virulence, motility, antibiotic production, and biofilm formation. Autoinducers come in a number of different forms depending on the species, but the effect that they have is similar in many cases. Autoinducers allow bacteria to communicate both within and between different species. This communication alters gene expression and allows bacteria to mount coordinated responses to their environments, in a manner that is comparable to behavior and signaling in higher organisms. Not surprisingly, it has been suggested that quorum sensing may have been an important evolutionary milestone that ultimately gave rise to multicellular life forms.

Virulence-related outer membrane proteins, or outer surface proteins (Osp) in some contexts, are expressed in the outer membrane of gram-negative bacteria and are essential to bacterial survival within macrophages and for eukaryotic cell invasion.

L-form bacteria, also known as L-phase bacteria, L-phase variants or cell wall-deficient (CWD) bacteria, are growth forms derived from different bacteria. They lack cell walls. Two types of L-forms are distinguished: unstable L-forms, spheroplasts that are capable of dividing, but can revert to the original morphology, and stable L-forms, L-forms that are unable to revert to the original bacteria.
Motility protein B also known as MotB is a bacterial protein that is encoded by the motB gene. It's a component of the flagellar motor. More specifically, MotA and MotB makes the stator of a flagellum and surround the rotor as a ring of about 8-10 particles. MotA and MotB are integral membrane proteins. While both MotA and MotB surround the MS ring, MotB also anchors MotA to cell wall peptidoglycan. These two proteins form pores that harvest energy for flagellar mechanical movement by proton motive force (PMF) across the membrane. Cellular metabolic processes such as the electron transport chain move protons outside the cell, creating more protons and more positive charge in the extracellular space. When the protons flow back into the cell through MotA and MotB along concentration and charge gradients, they release energy that is used for flagellar rotation. The speed of the flagellar motor is dependent on the magnitude of the PMF acting on MotA and MotB.

In molecular biology, YadA is a protein domain which is short for Yersinia adhesin A. These proteins have strong sequence and structural homology, particularly at their C-terminal end. The function is to promote their pathogenicity and virulence in host cells, though cell adhesion. YadA is found in three pathogenic species of Yersinia, Y. pestis,Y. pseudotuberculosis, and Y. enterocolitica. The YadA domain is encoded for by a virulence plasmid in Yersinia, which encodes a type-III secretion (T3S) system consisting of the Ysc injectisome and the Yop effectors.

Gemmata obscuriglobus is a species of Gram-negative, aerobic, heterotrophic bacteria of the phylum Planctomycetota. G. obscuriglobus occur in freshwater habitats and was first described in 1984, and is the only described species in its genus.