Related Research Articles

The zebrafish is a freshwater fish belonging to the minnow family (Cyprinidae) of the order Cypriniformes. Native to South Asia, it is a popular aquarium fish, frequently sold under the trade name zebra danio.

Xenopus is a genus of highly aquatic frogs native to sub-Saharan Africa. Twenty species are currently described within it. The two best-known species of this genus are Xenopus laevis and Xenopus tropicalis, which are commonly studied as model organisms for developmental biology, cell biology, toxicology, neuroscience and for modelling human disease and birth defects.
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, research, and forensics. Commonly made in the laboratory by solid-phase chemical synthesis, these small bits of nucleic acids can be manufactured as single-stranded molecules with any user-specified sequence, and so are vital for artificial gene synthesis, polymerase chain reaction (PCR), DNA sequencing, molecular cloning and as molecular probes. In nature, oligonucleotides are usually found as small RNA molecules that function in the regulation of gene expression, or are degradation intermediates derived from the breakdown of larger nucleic acid molecules.
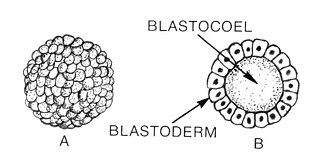
Blastulation is the stage in early animal embryonic development that produces the blastula. The blastula (from Greek βλαστός is a hollow sphere of cells surrounding an inner fluid-filled cavity. Embryonic development begins with a sperm fertilizing an egg cell to become a zygote, which undergoes many cleavages to develop into a ball of cells called a morula. Only when the blastocoel is formed does the early embryo become a blastula. The blastula precedes the formation of the gastrula in which the germ layers of the embryo form.
Gene knockdown is an experimental technique by which the expression of one or more of an organism's genes is reduced. The reduction can occur either through genetic modification or by treatment with a reagent such as a short DNA or RNA oligonucleotide that has a sequence complementary to either gene or an mRNA transcript.

A Morpholino, also known as a Morpholino oligomer and as a phosphorodiamidate Morpholino oligomer (PMO), is a type of oligomer molecule used in molecular biology to modify gene expression. Its molecular structure contains DNA bases attached to a backbone of methylenemorpholine rings linked through phosphorodiamidate groups. Morpholinos block access of other molecules to small specific sequences of the base-pairing surfaces of ribonucleic acid (RNA). Morpholinos are used as research tools for reverse genetics by knocking down gene function.

Spatiotemporal gene expression is the activation of genes within specific tissues of an organism at specific times during development. Gene activation patterns vary widely in complexity. Some are straightforward and static, such as the pattern of tubulin, which is expressed in all cells at all times in life. Some, on the other hand, are extraordinarily intricate and difficult to predict and model, with expression fluctuating wildly from minute to minute or from cell to cell. Spatiotemporal variation plays a key role in generating the diversity of cell types found in developed organisms; since the identity of a cell is specified by the collection of genes actively expressed within that cell, if gene expression was uniform spatially and temporally, there could be at most one kind of cell.

Zinc finger protein GLI2 also known as GLI family zinc finger 2 is a protein that in humans is encoded by the GLI2 gene. The protein encoded by this gene is a transcription factor.
Chordin or CHRD is a protein with a prominent role in dorsal–ventral patterning during vertebrate early embryonic development. Chordin is encoded by the chrd gene.
In developmental biology, midblastula or midblastula transition (MBT) occurs during the blastula stage of embryonic development. During this stage, the embryo is referred to as a blastula. The series of changes to the blastula that characterize the midblastula transition include activation of zygotic gene transcription, slowing of the cell cycle, increased asynchrony in cell division, and an increase in cell motility.

Growth differentiation factor 6 (GDF6) is a protein that in humans is encoded by the GDF6 gene.

Homeobox protein goosecoid is a protein that in humans is encoded by the GSC gene. This gene encodes a member of the bicoid subfamily of the paired (PRD) homeobox family of proteins. The encoded protein acts as a transcription factor and may be autoregulatory. A similar protein in mice plays a role in craniofacial and rib cage development during embryogenesis.
Xenbase is a Model Organism Database (MOD), providing informatics resources, as well as genomic and biological data on Xenopus frogs. Xenbase has been available since 1999, and covers both X. laevis and X. tropicalis Xenopus varieties. As of 2013 all of its services are running on virtual machines in a private cloud environment, making it one of the first MODs to do so. Other than hosting genomics data and tools, Xenbase supports the Xenopus research community though profiles for researchers and laboratories, and job and events postings.
Symmetry breaking in biology is the process by which uniformity is broken, or the number of points to view invariance are reduced, to generate a more structured and improbable state. That is to say, symmetry breaking is the event where symmetry along a particular axis is lost to establish a polarity. Polarity is a measure for a biological system to distinguish poles along an axis. This measure is important because it is the first step to building complexity. For example, during organismal development, one of the first steps for the embryo is to distinguish its dorsal-ventral axis. The symmetry-breaking event that occurs here will determine which end of this axis will be the ventral side, and which end will be the dorsal side. Once this distinction is made, then all the structures that are located along this axis can develop at the proper location. As an example, during human development, the embryo needs to establish where is ‘back’ and where is ‘front’ before complex structures, such as the spine and lungs, can develop in the right location. This relationship between symmetry breaking and complexity was articulated by P.W. Anderson. He speculated that increasing levels of broken symmetry in many-body systems correlates with increasing complexity and functional specialization. In a biological perspective, the more complex an organism is, the higher number of symmetry-breaking events can be found. Without symmetry breaking, building complexity in organisms would be very difficult.
The nodal signaling pathway is a signal transduction pathway important in regional and cellular differentiation during embryonic development.

In Xenopus laevis, the specification of the three germ layers occurs at the blastula stage. Great efforts have been made to determine the factors that specify the endoderm and mesoderm. On the other hand, only a few examples of genes that are required for ectoderm specification have been described in the last decade. The first molecule identified to be required for the specification of ectoderm was the ubiquitin ligase Ectodermin ; later, it was found that the deubiquitinating enzyme, FAM/USP9x, is able to overcome the effects of ubiquitination made by Ectodermin in Smad4. Two transcription factors have been proposed to control gene expression of ectodermal specific genes: POU91/Oct3/4 and FoxIe1/Xema. A new factor specific for the ectoderm, XFDL156, has shown to be essential for suppression of mesoderm differentiation from pluripotent cells.
This article is about the role of Fibroblast Growth Factor Signaling in Mesoderm Formation.
Maternal to zygotic transition is the stage in embryonic development during which development comes under the exclusive control of the zygotic genome rather than the maternal (egg) genome. The egg contains stored maternal genetic material mRNA which controls embryo development until the onset of MZT. After MZT the diploid embryo takes over genetic control. This requires both zygotic genome activation (ZGA) and degradation of maternal products. This process is important because it is the first time that the new embryonic genome is utilized and the paternal and maternal genomes are used in combination. The zygotic genome now drives embryo development.
Chemical genetics is the investigation of the function of proteins and signal transduction pathways in cells by the screening of chemical libraries of small molecules. Chemical genetics is analogous to classical genetic screen where random mutations are introduced in organisms, the phenotype of these mutants is observed, and finally the specific gene mutation (genotype) that produced that phenotype is identified. In chemical genetics, the phenotype is disturbed not by introduction of mutations, but by exposure to small molecule tool compounds. Phenotypic screening of chemical libraries is used to identify drug targets or to validate those targets in experimental models of disease. Recent applications of this topic have been implicated in signal transduction, which may play a role in discovering new cancer treatments. Chemical genetics can serve as a unifying study between chemistry and biology. The approach was first proposed by Tim Mitchison in 1994 in an opinion piece in the journal Chemistry & Biology entitled "Towards a pharmacological genetics".
Reverse genetics is a method in molecular genetics that is used to help understand the function(s) of a gene by analysing the phenotypic effects caused by genetically engineering specific nucleic acid sequences within the gene. The process proceeds in the opposite direction to forward genetic screens of classical genetics. While forward genetics seeks to find the genetic basis of a phenotype or trait, reverse genetics seeks to find what phenotypes are controlled by particular genetic sequences.
References
- ↑ Ekker SC (December 2000). "Morphants: a new systematic vertebrate functional genomics approach". Yeast. 17 (4): 302–306. doi:10.1002/1097-0061(200012)17:4<302::AID-YEA53>3.0.CO;2-#. PMC 2448384 . PMID 11119307.
- ↑ Nasevicius A, Ekker SC (October 2000). "Effective targeted gene 'knockdown' in zebrafish". Nat. Genet. 26 (2): 216–20. doi:10.1038/79951. PMID 11017081. S2CID 21451111.
- ↑ Heasman J, Kofron M, Wylie C (June 2000). "Beta-catenin signaling activity dissected in the early Xenopus embryo: a novel antisense approach". Dev. Biol. 222 (1): 124–34. doi:10.1006/dbio.2000.9720. PMID 10885751.