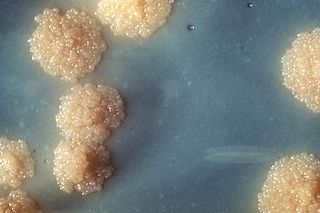
Mycobacterium tuberculosis, also known as Koch's bacillus, is a species of pathogenic bacteria in the family Mycobacteriaceae and the causative agent of tuberculosis. First discovered in 1882 by Robert Koch, M. tuberculosis has an unusual, waxy coating on its cell surface primarily due to the presence of mycolic acid. This coating makes the cells impervious to Gram staining, and as a result, M. tuberculosis can appear weakly Gram-positive. Acid-fast stains such as Ziehl–Neelsen, or fluorescent stains such as auramine are used instead to identify M. tuberculosis with a microscope. The physiology of M. tuberculosis is highly aerobic and requires high levels of oxygen. Primarily a pathogen of the mammalian respiratory system, it infects the lungs. The most frequently used diagnostic methods for tuberculosis are the tuberculin skin test, acid-fast stain, culture, and polymerase chain reaction.

Mycobacterium is a genus of over 190 species in the phylum Actinomycetota, assigned its own family, Mycobacteriaceae. This genus includes pathogens known to cause serious diseases in mammals, including tuberculosis and leprosy in humans. The Greek prefix myco- means 'fungus', alluding to this genus' mold-like colony surfaces. Since this genus has cell walls with a waxy lipid-rich outer layer that contains high concentrations of mycolic acid, acid-fast staining is used to emphasize their resistance to acids, compared to other cell types.
Nontuberculous mycobacteria (NTM), also known as environmental mycobacteria, atypical mycobacteria and mycobacteria other than tuberculosis (MOTT), are mycobacteria which do not cause tuberculosis or leprosy/Hansen's disease. NTM are able to cause pulmonary diseases that resemble tuberculosis. Mycobacteriosis is any of these illnesses, usually meant to exclude tuberculosis. They occur in many animals, including humans and are commonly found in soil and water.

Mycobacterium smegmatis is an acid-fast bacterial species in the phylum Actinomycetota and the genus Mycobacterium. It is 3.0 to 5.0 μm long with a bacillus shape and can be stained by Ziehl–Neelsen method and the auramine-rhodamine fluorescent method. It was first reported in November 1884 by Lustgarten, who found a bacillus with the staining appearance of tubercle bacilli in syphilitic chancres. Subsequent to this, Alvarez and Tavel found organisms similar to that described by Lustgarten also in normal genital secretions (smegma). This organism was later named M. smegmatis.
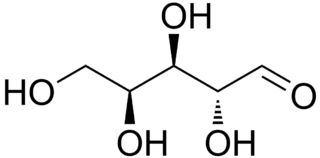
Lyxose is an aldopentose — a monosaccharide containing five carbon atoms, and including an aldehyde functional group. It has chemical formula C5H10O5. It is a C'-2 carbon epimer of the sugar xylose. The name "lyxose" comes from reversing the prefix "xyl" in "xylose".
Mycolic acids are long fatty acids found in the cell walls of Mycobacteriales taxon, a group of bacteria that includes Mycobacterium tuberculosis, the causative agent of the disease tuberculosis. They form the major component of the cell wall of many Mycobacteriales species. Despite their name, mycolic acids have no biological link to fungi; the name arises from the filamentous appearance their presence gives Mycobacteriales under high magnification. The presence of mycolic acids in the cell wall also gives Mycobacteriales a distinct gross morphological trait known as "cording". Mycolic acids were first isolated by Stodola et al. in 1938 from an extract of M. tuberculosis.

A mycobacteriophage is a member of a group of bacteriophages known to have mycobacteria as host bacterial species. While originally isolated from the bacterial species Mycobacterium smegmatis and Mycobacterium tuberculosis, the causative agent of tuberculosis, more than 4,200 mycobacteriophage have since been isolated from various environmental and clinical sources. 2,042 have been completely sequenced. Mycobacteriophages have served as examples of viral lysogeny and of the divergent morphology and genetic arrangement characteristic of many phage types.
The Timpe and Runyon classification of nontuberculous mycobacteria based on the rate of growth, production of yellow pigment and whether this pigment was produced in the dark or only after exposure to light.
Lipoarabinomannan, also called LAM, is a glycolipid, and a virulence factor associated with Mycobacterium tuberculosis, the bacteria responsible for tuberculosis. Its primary function is to inactivate macrophages and scavenge oxidative radicals.
Mycobacterium caprae is a species of bacteria in the genus Mycobacterium and a member of the Mycobacterium tuberculosis complex. The species is named after the caprines, the organisms from which M. caprae was first isolated. Prior to 2003, the species was referred to as Mycobacterium tuberculosis subsp. caprae. It is also synonymous with the name Mycobacterium bovis subsp. caprae.

Mycobacterium tuberculosis contains at least nine small RNA families in its genome. The small RNA (sRNA) families were identified through RNomics – the direct analysis of RNA molecules isolated from cultures of Mycobacterium tuberculosis. The sRNAs were characterised through RACE mapping and Northern blot experiments. Secondary structures of the sRNAs were predicted using Mfold.
Giles is a bacteriophage that infects Mycobacterium smegmatis bacteria. The genome of this phage is very different from that of other mycobacteriophages and is highly mosaic. More than half of its predicted genes are novel and are not seen in other species.
Glucose-6-phosphate dehydrogenase (coenzyme-F420) is an enzyme with systematic name D-glucose-6-phosphate:F420 1-oxidoreductase. This enzyme catalyses the following chemical reaction
UDP-N-acetylglucosamine---decaprenyl-phosphate N-acetylglucosaminephosphotransferase is an enzyme with systematic name UDP-N-acetyl-alpha-D-glucosamine:trans,octacis-decaprenyl-phosphate N-acetylglucosaminephosphotransferase. This enzyme catalyses the following chemical reaction
Mycobacterium virus Patience, also called Patience, is a bacteriophage that infects Mycobacterium smegmatis bacteria. The large difference between the GC content of this virus's genome (50.3%) and that of its host (67.4%) indicate Patience likely evolved among bacteria of lower GC content but was able to infect M. smegmatis as well. It is the only species of the genus Patiencevirus.
The Mycobacterial 2 TMS Phage Holin Family is a group of transporters belonging to the Holin Superfamily VII. The Mycobactrerial 2 transmembrane segment (TMS) Holins have been identified and recognized by Catalao et al (2012). The Mycobacterium phage D29 gp11 protein is a holin that, upon expression, rapidly kills both E. coli and Mycobacterium smegmatis. Shortening gp11 from its C-terminus resulted in diminished cytotoxicity and smaller holes. The two TMSs at the N-terminus alone do not integrate into the cytoplasmic membrane and do not show toxicity. Fusion of the two TMSs and a small C-terminal coiled-coil region resulted in restoration of cell killing. The second TMS is dispensable for toxicity. The gp11 C-terminal region is therefore necessary but not sufficient for toxicity.
Tathamangalam Ananthanarayanan Venkitasubramanian (1924–2003), popularly known as TAV, was an Indian biochemist, known for his researches on tuberculosis and the biochemistry of bacillus. He was a professor and the head of the department of biochemistry at Vallabhbhai Patel Chest Institute, Delhi. The Council of Scientific and Industrial Research, the apex agency of the Government of India for scientific research, awarded him the Shanti Swarup Bhatnagar Prize for Science and Technology, one of the highest Indian science awards, in 1968, for his contributions to biological sciences.
Mycobacterium virus Jeffabunny is a bacteriophage known to infect bacterial species of the genus Mycobacterium. It was discovered by Jeffrey Rubin in 2008 in a Howard Hughes Medical Institute funded lab as part of SEA-PHAGES. It was taken from surface soil in a planter, in the shade of a non-redwood tree at Merrill College found infecting, specifically, Mycobacterium smegmatis.
Mycobacterium virus D29 (D29) is a cluster A mycobacteriophage belonging to the Siphoviridae family of viruses, it was discovered in 1954 by S. Froman. D29 is notable for its ability to infect M. tuberculosis. D29 is a double stranded DNA mycobacteriophage. It is a lytic phage, this means that D29 takes the lytic pathway of infection instead of the lysogenic pathway of infection. There are no human associated diseases associated with mycobacterium virus D29.
Unicornvirus is a genus of bacteriophage viruses in the class Caudoviricetes. The genus was named for the first virus of this type, Unicornvirus unicorn, which was originally called "Mycobacterium phage Unicorn".





