| Mycobacterium | |
|---|---|
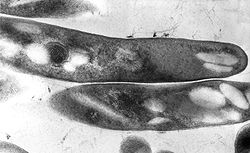 | |
| TEM micrograph of M. tuberculosis | |
| Scientific classification | |
| Domain: | Bacteria |
| Kingdom: | Bacillati |
| Phylum: | Actinomycetota |
| Class: | Actinomycetes |
| Order: | Mycobacteriales |
| Family: | Mycobacteriaceae |
| Genus: | Mycobacterium Lehmann & Neumann 1896 [1] |
| Species | |
Over 190 species, see LPSN | |
| Synonyms [2] | |
| |
Mycobacterium is a genus of over 190 species of Gram-positive bacteria in the phylum Actinomycetota, assigned its own family, Mycobacteriaceae. This genus includes pathogens known to cause serious diseases in mammals, including tuberculosis ( M. tuberculosis ) and leprosy ( M. leprae ) in humans. The Greek prefix myco- means 'fungus', alluding to this genus's mold-like colony surfaces. [3] Since this genus has cell walls with a waxy lipid-rich outer layer containing high concentrations of mycolic acid, [4] acid-fast staining is used to emphasize their resistance to acids, compared to other cell types. [5]
Contents
- Microbiology
- Morphology
- Physiology
- Ecology
- Genomics
- Pathogenicity
- Mycobacterium tuberculosis complex
- Leprosy
- Nontuberculosis Mycobacteria
- Virulence factors
- History
- Proposed division of the genus
- Diagnosis
- Mycobacteriophages
- Mycosides
- Notes
- References
- External links
Mycobacterial species are generally aerobic, non-motile, and capable of growing with minimal nutrition. The genus is divided based on each species's pigment production and growth rate. [6] While most Mycobacterium species are non-pathogenic, the genus's characteristic complex cell wall contributes to evasion from host defenses. [7]






