Related Research Articles

Mycobacterium is a genus of Actinomycetota, given its own family, the Mycobacteriaceae. Over 190 species are recognized in this genus. This genus includes pathogens known to cause serious diseases in mammals, including tuberculosis and leprosy in humans. The Greek prefix myco- means 'fungus', alluding to the way mycobacteria have been observed to grow in a mold-like fashion on the surface of cultures. It is acid-fast and cannot be stained by the Gram stain procedure.
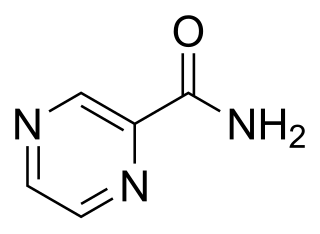
Pyrazinamide is a medication used to treat tuberculosis. For active tuberculosis, it is often used with rifampicin, isoniazid, and either streptomycin or ethambutol. It is not generally recommended for the treatment of latent tuberculosis. It is taken by mouth.
Mycobacterium botniense is a slowly growing Mycobacterium, which produces a yellow pigment. It was first isolated from a stream of water. M. botniense is most closely related to Mycobacterium xenopi. Etymology: botniense; of Botnia, referring to the Latin name of the province of Finland from which the isolation was made.
Mycobacterium branderi is a slowly growing, nonchromogenic Mycobacterium first isolated from patients in Finland. Etymology: of Brander, referring to Eljas Brander, the former head of the Tuberculosis Laboratory of the National Public Health Institute, Finland, who collected the strains.
Mycobacterium brisbanense is a member of the Mycobacterium fortuitum third biovariant complex. They are rapidly growing ubiquitous environmental organisms that normally inhabit soil, dust and water. These organisms frequently are human pathogens that cause a wide spectrum of clinically significant disease. It is important for practitioners to be aware of these organisms as possible etiological agents, as they are resistant to most first-line anti-tuberculous agents.
Mycobacterium brumae is a rapidly growing environmental mycobacterial species identified in 1993. Aside from one 2004 report of a catheter related bloodstream infection no other infections by this organism have been reported. It was first isolated from water, soil and one human sputum sample in Spain.
Mycobacterium canariasense is a rapidly growing, non-pigmented mycobacterium first isolated from blood samples obtained from 17 patients with febrile syndrome. Etymology: canariasense; referring to the Latin adjective of the Spanish islands where all strains were isolated.
Mycobacterium conspicuum is a species of the phylum Actinomycetota, belonging to the genus Mycobacterium.

Mycobacterium cosmeticum is a rapidly growing mycobacterium that was first isolated from cosmetic patients and sites performing cosmetic procedures.
Mycobacterium elephantis, a bacterium of the family Mycobacteriaceae, was discovered and isolated from a deceased elephant near India and may be linked to respiratory dysfunction. Organisms in the genus Mycobacterium are known to be aerobic and non-motile. Organisms within Mycobacterium belong to either the rapid growing group or the slow growing group. M. elephantis is classified as a rapid grower and relates most closely to Mycobacterium confluentis and Mycobacterium phlei.
Mycobacterium fallax is a species of the phylum Actinomycetota, belonging to the genus Mycobacterium.
Mycobacterium gordonae is a species of Mycobacterium named for Ruth E. Gordon. It is a species of the phylum Actinomycetota, belonging to the genus Mycobacterium.
Mycobacterium hassiacum is a rapid-growing thermophilic mycobacterium that was isolated in human urine in 1997 by researchers at the German University of Regensburg. It's a species of the phylum Actinomycetota, belonging to the genus Mycobacterium.

Mycobacterium kansasii is a bacterium in the Mycobacterium genus. It is an environmental bacteria that causes opportunistic infections in humans, and is the one of the leading mycobacterial causes of human disease after tuberculosis and leprosy.
Mycobacterium wolinskyi is a rapidly growing mycobacterium most commonly seen in post-traumatic wound infections, especially those following open fractures and with associated osteomyelitis. Mycobacterium wolinskyi is clearly clinically significant, and occurs in the same settings as Mycobacterium smegmatis and members of the Mycobacterium fortuitum complex; they differ from members of the Mycobacterium fortuitum complex in the type of chronic lung disease they produce, with essentially all cases occurring in the setting of chronic lipoid pneumonia, either secondary to chronic oil ingestion or chronic aspiration. Etymology: Wolinsky, named after Emanuel Wolinsky in honour of, and in recognition for, significant contributions to the study of the non-tuberculous mycobacteria.

Löwenstein–Jensen medium, more commonly known as LJ medium, is a growth medium specially used for culture of Mycobacterium species, notably Mycobacterium tuberculosis.
Neisseria cinerea is a commensal species grouped with the Gram-negative, oxidase-positive, and catalase-positive diplococci. It was first classified as Micrococcus cinereus by Alexander von Lingelsheim in 1906. Using DNA hybridization, N. cinerea exhibits 50% similarity to Neisseria gonorrhoeae.
In microbiology, the term isolation refers to the separation of a strain from a natural, mixed population of living microbes, as present in the environment, for example in water or soil, or from living beings with skin flora, oral flora or gut flora, in order to identify the microbe(s) of interest. Historically, the laboratory techniques of isolation first developed in the field of bacteriology and parasitology, before those in virology during the 20th century.

The niacin test has been widely used since the 1960s to identify mycobacteria at the species level in the clinical laboratory. The niacin test detects niacin in aqueous extracts of a culture. M. tuberculosis strains that test negative for the niacin test are very rare. Redox reactions happening in Mycobacterium species produce niacin as a part of energy metabolism. Even though all mycobacteria produce niacin, M. tuberculosis accumulates an excess of niacin because of its inability to process niacin, excreting the excess niacin into the culture media, thus allowing it to be detected using the niacin test. The niacin test is typically only conducted on slow-growing, granular, tan colored colonies, as these are the morphology characteristics of M. tuberculosis on an agar plate. Because of its affordability compared to expensive identification methods like pyrosequencing or MALDI-TOF MS that require expensive machines and reagents.
Diagnostic microbiology is the study of microbial identification. Since the discovery of the germ theory of disease, scientists have been finding ways to harvest specific organisms. Using methods such as differential media or genome sequencing, physicians and scientists can observe novel functions in organisms for more effective and accurate diagnosis of organisms. Methods used in diagnostic microbiology are often used to take advantage of a particular difference in organisms and attain information about what species it can be identified as, which is often through a reference of previous studies. New studies provide information that others can reference so that scientists can attain a basic understanding of the organism they are examining.
References
- 1 2 3 4 5 6 7 8 9 10 11 12 13 Koneman, E. (1988). Diagnostic Microbiology. Philadelphia: J.B. Lippincott.
- ↑ ACHARYA, TANKESHWAR. "Key biochemical methods used to distinguish Mycobacterial group" . Retrieved 21 November 2014.
- 1 2 3 Metchock, B.G; Nolte, F.S.; Wallace, R.J. (1999). "Mycobacterium". In Murray, P.R.; Baron, E.J.; Pfaller, M.A.; Tenover, F.C.; Yolken, R.H. (eds.). Manual of Clinical Microbiology. Washington, D.C.: ASM Press. pp. 399–427.
- ↑ Deutsches Institut für Normung (1993). "Part 9: minimum requirements for the identification of tubercle bacilli". Medical Microbiology: Diagnosis of Tuberculosis. Berlin: Beuth Verlag (DIN 58943-9).
- ↑ M. Tsukamura. "Adansonian classification of mycobacteria". Journal of General Microbiology. 45: 252–273. doi: 10.1099/00221287-45-2-253 .