Related Research Articles
An electron transport chain (ETC) is a series of protein complexes and other molecules which transfer electrons from electron donors to electron acceptors via redox reactions (both reduction and oxidation occurring simultaneously) and couples this electron transfer with the transfer of protons (H+ ions) across a membrane. Many of the enzymes in the electron transport chain are embedded within the membrane.
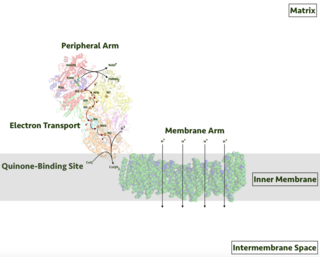
Respiratory complex I, EC 7.1.1.2 is the first large protein complex of the respiratory chains of many organisms from bacteria to humans. It catalyzes the transfer of electrons from NADH to coenzyme Q10 (CoQ10) and translocates protons across the inner mitochondrial membrane in eukaryotes or the plasma membrane of bacteria.

Nicotinamide adenine dinucleotide (NAD) is a coenzyme central to metabolism. Found in all living cells, NAD is called a dinucleotide because it consists of two nucleotides joined through their phosphate groups. One nucleotide contains an adenine nucleobase and the other, nicotinamide. NAD exists in two forms: an oxidized and reduced form, abbreviated as NAD+ and NADH (H for hydrogen), respectively.

In biochemistry, flavin adenine dinucleotide (FAD) is a redox-active coenzyme associated with various proteins, which is involved with several enzymatic reactions in metabolism. A flavoprotein is a protein that contains a flavin group, which may be in the form of FAD or flavin mononucleotide (FMN). Many flavoproteins are known: components of the succinate dehydrogenase complex, α-ketoglutarate dehydrogenase, and a component of the pyruvate dehydrogenase complex.

Flavin mononucleotide (FMN), or riboflavin-5′-phosphate, is a biomolecule produced from riboflavin (vitamin B2) by the enzyme riboflavin kinase and functions as the prosthetic group of various oxidoreductases, including NADH dehydrogenase, as well as a cofactor in biological blue-light photo receptors. During the catalytic cycle, a reversible interconversion of the oxidized (FMN), semiquinone (FMNH•), and reduced (FMNH2) forms occurs in the various oxidoreductases. FMN is a stronger oxidizing agent than NAD and is particularly useful because it can take part in both one- and two-electron transfers. In its role as blue-light photo receptor, (oxidized) FMN stands out from the 'conventional' photo receptors as the signaling state and not an E/Z isomerization.

Flavoproteins are proteins that contain a nucleic acid derivative of riboflavin. These proteins are involved in a wide array of biological processes, including removal of radicals contributing to oxidative stress, photosynthesis, and DNA repair. The flavoproteins are some of the most-studied families of enzymes.
Any enzyme system that includes cytochrome P450 protein or domain can be called a P450-containing system.

Nitrate reductases are molybdoenzymes that reduce nitrate to nitrite. This reaction is critical for the production of protein in most crop plants, as nitrate is the predominant source of nitrogen in fertilized soils.

In enzymology, a D-xylulose reductase (EC 1.1.1.9) is an enzyme that is classified as an Oxidoreductase (EC 1) specifically acting on the CH-OH group of donors (EC 1.1.1) that uses NAD+ or NADP+ as an acceptor (EC 1.1.1.9). This enzyme participates in pentose and glucuronate interconversions; a set of metabolic pathways that involve converting pentose sugars and glucuronate into other compounds.
In enzymology, a ferric-chelate reductase (EC 1.16.1.7) is an enzyme that catalyzes the chemical reaction

In enzymology, a NADH peroxidase (EC 1.11.1.1) is an enzyme that catalyzes the chemical reaction
Flavin reductase a class of enzymes. There are a variety of flavin reductases, which bind free flavins and through hydrogen bonding, catalyze the reduction of these molecules to a reduced flavin. Riboflavin, or vitamin B, and flavin mononucleotide are two of the most well known flavins in the body and are used in a variety of processes which include metabolism of fat and ketones and the reduction of methemoglobin in erythrocytes. Flavin reductases are similar and often confused for ferric reductases because of their similar catalytic mechanism and structures.
In enzymology, an FMN reductase (EC 1.5.1.29) is an enzyme that catalyzes the chemical reaction
In enzymology, a NAD(P)H dehydrogenase (quinone) (EC 1.6.5.2) is an enzyme that catalyzes the chemical reaction

In enzymology, a pyrroline-5-carboxylate reductase (EC 1.5.1.2) is an enzyme that catalyzes the chemical reaction
Deiodinase (monodeiodinase) is a peroxidase enzyme that is involved in the activation or deactivation of thyroid hormones.
FAD reductase (NADH) (EC 1.5.1.37, NADH-FAD reductase, NADH-dependent FAD reductase) is an enzyme with systematic name FADH2:NAD+ oxidoreductase. This enzyme catalyses the following chemical reaction
FMN reductase (NAD(P)H) (EC 1.5.1.39, FRG) is an enzyme with systematic name FMNH2:NAD(P)+ oxidoreductase. This enzyme catalyses the following chemical reaction
Riboflavin reductase (NAD(P)H) (EC 1.5.1.41, NAD(P)H-FMN reductase, Fre) is an enzyme with systematic name riboflavin:NAD(P)+ oxidoreductase. This enzyme catalyses the following chemical reaction
FMN reductase (NADH) (EC 1.5.1.42, NADH-FMN reductase) is an enzyme with systematic name FMNH2:NAD+ oxidoreductase. This enzyme catalyses the following chemical reaction
References
- ↑ Hecht HJ, Erdmann H, Park HJ, Sprinzl M, Schmid RD (December 1995). "Crystal structure of NADH oxidase from Thermus thermophilus". Nat. Struct. Biol. 2 (12): 1109–14. doi:10.1038/nsb1295-1109. PMID 8846223. S2CID 8384273.
- ↑ de Oliveira IM, Henriques JA, Bonatto D (April 2007). "In silico identification of a new group of specific bacterial and fungal nitroreductases-like proteins". Biochem. Biophys. Res. Commun. 355 (4): 919–25. doi:10.1016/j.bbrc.2007.02.049. PMID 17331467.
- ↑ Bryant, C.; DeLuca, M. (1991-03-05). "Purification and characterization of an oxygen-insensitive NAD(P)H nitroreductase from Enterobacter cloacae". Journal of Biological Chemistry. 266 (7): 4119–4125. ISSN 0021-9258. PMID 1999405.
- ↑ Çelik, Ayhan; Yetiş, Gülden (2012-06-01). "An unusually cold active nitroreductase for prodrug activations". Bioorganic & Medicinal Chemistry. 20 (11): 3540–3550. doi:10.1016/j.bmc.2012.04.004. PMID 22546205.