
In molecular biology, RNA polymerase, or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that catalyzes the chemical reactions that synthesize RNA from a DNA template.
In molecular biology and genetics, transcriptional regulation is the means by which a cell regulates the conversion of DNA to RNA (transcription), thereby orchestrating gene activity. A single gene can be regulated in a range of ways, from altering the number of copies of RNA that are transcribed, to the temporal control of when the gene is transcribed. This control allows the cell or organism to respond to a variety of intra- and extracellular signals and thus mount a response. Some examples of this include producing the mRNA that encode enzymes to adapt to a change in a food source, producing the gene products involved in cell cycle specific activities, and producing the gene products responsible for cellular differentiation in multicellular eukaryotes, as studied in evolutionary developmental biology.
A sigma factor is a protein needed for initiation of transcription in bacteria. It is a bacterial transcription initiation factor that enables specific binding of RNA polymerase (RNAP) to gene promoters. It is homologous to archaeal transcription factor B and to eukaryotic factor TFIIB. The specific sigma factor used to initiate transcription of a given gene will vary, depending on the gene and on the environmental signals needed to initiate transcription of that gene. Selection of promoters by RNA polymerase is dependent on the sigma factor that associates with it. They are also found in plant chloroplasts as a part of the bacteria-like plastid-encoded polymerase (PEP).

The preinitiation complex is a complex of approximately 100 proteins that is necessary for the transcription of protein-coding genes in eukaryotes and archaea. The preinitiation complex positions RNA polymerase II at gene transcription start sites, denatures the DNA, and positions the DNA in the RNA polymerase II active site for transcription.

RNA polymerase II is a multiprotein complex that transcribes DNA into precursors of messenger RNA (mRNA) and most small nuclear RNA (snRNA) and microRNA. It is one of the three RNAP enzymes found in the nucleus of eukaryotic cells. A 550 kDa complex of 12 subunits, RNAP II is the most studied type of RNA polymerase. A wide range of transcription factors are required for it to bind to upstream gene promoters and begin transcription.

General transcription factors (GTFs), also known as basal transcriptional factors, are a class of protein transcription factors that bind to specific sites (promoter) on DNA to activate transcription of genetic information from DNA to messenger RNA. GTFs, RNA polymerase, and the mediator constitute the basic transcriptional apparatus that first bind to the promoter, then start transcription. GTFs are also intimately involved in the process of gene regulation, and most are required for life.
In eukaryote cells, RNA polymerase III is a protein that transcribes DNA to synthesize 5S ribosomal RNA, tRNA and other small RNAs.

Eukaryotic transcription is the elaborate process that eukaryotic cells use to copy genetic information stored in DNA into units of transportable complementary RNA replica. Gene transcription occurs in both eukaryotic and prokaryotic cells. Unlike prokaryotic RNA polymerase that initiates the transcription of all different types of RNA, RNA polymerase in eukaryotes comes in three variations, each translating a different type of gene. A eukaryotic cell has a nucleus that separates the processes of transcription and translation. Eukaryotic transcription occurs within the nucleus where DNA is packaged into nucleosomes and higher order chromatin structures. The complexity of the eukaryotic genome necessitates a great variety and complexity of gene expression control.

Mediator is a multiprotein complex that functions as a transcriptional coactivator in all eukaryotes. It was discovered in 1990 in the lab of Roger D. Kornberg, recipient of the 2006 Nobel Prize in Chemistry. Mediator complexes interact with transcription factors and RNA polymerase II. The main function of mediator complexes is to transmit signals from the transcription factors to the polymerase.

The positive transcription elongation factor, P-TEFb, is a multiprotein complex that plays an essential role in the regulation of transcription by RNA polymerase II in eukaryotes. Immediately following initiation Pol II becomes trapped in promoter proximal paused positions on the majority of human genes. P-TEFb is a cyclin dependent kinase that can phosphorylate the DRB sensitivity inducing factor (DSIF) and negative elongation factor (NELF), as well as the carboxyl terminal domain of the large subunit of Pol II and this causes the transition into productive elongation leading to the synthesis of mRNAs. P-TEFb is regulated in part by a reversible association with the 7SK snRNP. Treatment of cells with the P-TEFb inhibitors DRB or flavopidirol leads to loss of mRNA production and ultimately cell death.
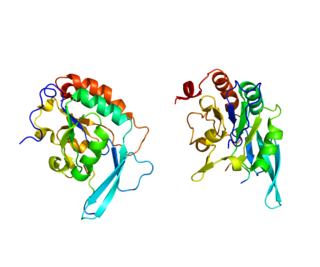
DNA-directed RNA polymerase II subunit RPB1, also known as RPB1, is an enzyme that is encoded by the POLR2A gene in humans.

Negative elongation factor E is a protein that in humans is encoded by the RDBP gene.

Cofactor of BRCA1, also known as COBRA1, is a human gene that encodes NELF-B.
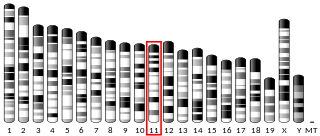
Transcription elongation factor SPT4 is a protein that in humans is encoded by the SUPT4H1 gene.

Negative elongation factor A is a protein that in humans is encoded by the WHSC2 gene.

Negative elongation factor C/D is a protein that in humans is encoded by the TH1L gene.

Mediator of RNA polymerase II transcription subunit 26 is an enzyme that in humans is encoded by the MED26 gene. It forms part of the Mediator complex.
RNA polymerase II holoenzyme is a form of eukaryotic RNA polymerase II that is recruited to the promoters of protein-coding genes in living cells. It consists of RNA polymerase II, a subset of general transcription factors, and regulatory proteins known as SRB proteins.
DSIF is a protein complex that can either negatively or positively affect transcription by RNA polymerase II. It can interact with the negative elongation factor (NELF) to promote the stalling of Pol II at some genes. This stalling is relieved by P-TEFb.

Mediator complex subunit 13 is a protein that in humans is encoded by the MED13 gene.
















