A transmembrane protein (TP) is a type of integral membrane protein that spans the entirety of the cell membrane to which it is permanently attached. Many transmembrane proteins function as gateways to permit the transport of specific substances across the membrane. They frequently undergo significant conformational changes to move a substance through the membrane.

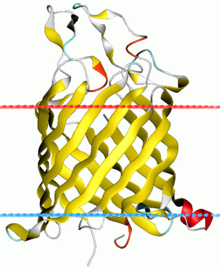
Porins are beta barrel proteins that cross a cellular membrane and act as a pore, through which molecules can diffuse. Unlike other membrane transport proteins, porins are large enough to allow passive diffusion, i.e., they act as channels that are specific to different types of molecules. They are present in the outer membrane of gram-negative bacteria and some gram-positive Mycobacteria, the outer membrane of mitochondria, and the outer chloroplast membrane.

The bacterial outer membrane is found in gram-negative bacteria. Its composition is distinct from that of the inner cytoplasmic cell membrane - among other things, the outer leaflet of the outer membrane of many gram-negative bacteria includes a complex lipopolysaccharide whose lipid portion acts as an endotoxin - and in some bacteria such as E. coli it is linked to the cell's peptidoglycan by Braun's lipoprotein.
The Transporter Classification Database is an International Union of Biochemistry and Molecular Biology (IUBMB)-approved classification system for membrane transport proteins, including ion channels.
Bacterial display is a protein engineering technique used for in vitro protein evolution. Libraries of polypeptides displayed on the surface of bacteria can be screened using flow cytometry or iterative selection procedures (biopanning). This protein engineering technique allows us to link the function of a protein with the gene that encodes it. Bacterial display can be used to find target proteins with desired properties and can be used to make affinity ligands which are cell-specific. This system can be used in many applications including the creation of novel vaccines, the identification of enzyme substrates and finding the affinity of a ligand for its target protein.
Virulence factors are molecules produced by bacteria, viruses, fungi, and protozoa that add to their effectiveness and enable them to achieve the following:
Protein K is a porin expressed in some pathogenic strains of E. coli bacteria. It has a molecular weight of about 40 kDa and is localized to the outer membrane, through which it allows both inorganic and organic ions to pass.

Intimin is a virulence factor (adhesin) of EPEC and EHEC E. coli strains. It is an attaching and effacing (A/E) protein, which with other virulence factors is necessary and responsible for enteropathogenic and enterohaemorrhagic diarrhoea.

Outer membrane receptors, also known as TonB-dependent receptors, are a family of beta barrel proteins named for their localization in the outer membrane of gram-negative bacteria. TonB complexes sense signals from the outside of bacterial cells and transmit them into the cytoplasm, leading to transcriptional activation of target genes. TonB-dependent receptors in gram-negative bacteria are associated with the uptake and transport of large substrates such as iron siderophore complexes and vitamin B12.
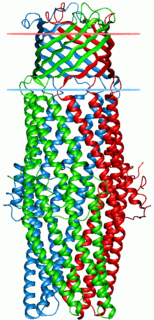
Proteins in the outer membrane efflux protein family form trimeric (three-piece) channels that allow export of a variety of substrates in gram-negative bacteria. Each member of this family is composed of two repeats. The trimeric channel is composed of a 12-stranded beta-barrel that spans the outer membrane, and a long all helical barrel that spans the periplasm.
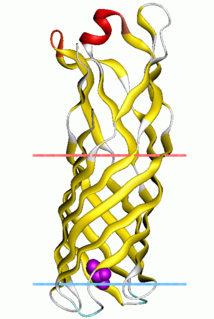
Outer membrane protein W (OmpW) family is a family of evolutionarily related proteins from the outer bacterial membrane.
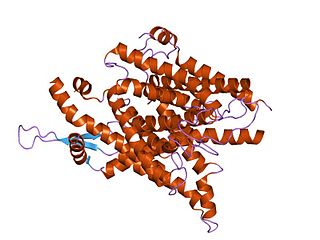
The SecY protein is the main transmembrane subunit of the bacterial Sec or Type II secretory pathway and a protein-secreting ATPase complex, also known as a SecYEG translocon. Homologs of the SecYEG complex are found in eukaryotes, where the subunit is known as Sec61α, and in archaea.

Outer membrane protein G (OmpG) is a porin, a channel proteins in the outer membrane of Gram-negative bacteria.
Omptins are a family of bacterial proteases. They are aspartate proteases, which cleave peptides with the use of a water molecule. Found in the outer membrane of gram-negative enterobacteria such as Shigella flexneri, Yersinia pestis, Escherichia coli, and Salmonella enterica. Omptins consist of a widely conserved beta barrel spanning the membrane with 5 extra-cellular loops. These loops are responsible for the various substrate specificities. These proteases rely upon binding of lipopolysaccharide for activity.
EnvZ/OmpR is a two-component regulatory system widely distributed in bacteria and particularly well characterized in Escherichia coli. Its function is in osmoregulation, responding to changes in environmental osmolality by regulating the expression of the outer membrane porins OmpF and OmpC. EnvZ is a histidine kinase which also possesses a cytoplasmic osmosensory domain, and OmpR is its corresponding response regulator protein.
The capsule biosynthesis, or CPS operon is a section of the genome present in some Escherichia coli which regulates the production of polysaccharides which make up the bacterial capsule. These polysaccharides help protect the bacteria from harsh environments, toxic chemicals, and bacteriophages.

In molecular biology, the OmpA domain is a conserved protein domain with a beta/alpha/beta/alpha-beta(2) structure found in the C-terminal region of many Gram-negative bacterial outer membrane proteins, such as porin-like integral membrane proteins, small lipid-anchored proteins, and MotB proton channels. The N-terminal half of these proteins is variable although some of the proteins in this group have the OmpA-like transmembrane domain at the N terminus. OmpA from Escherichia coli is required for pathogenesis, and can interact with host receptor molecules. MotB serve two functions in E. coli, the MotA(4)-MotB(2) complex attaches to the cell wall via MotB to form the stator of the flagellar motor, and the MotA-MotB complex couples the flow of ions across the cell membrane to movement of the rotor.
Phosphate-selective porins are a family of outer bacterial membrane proteins. These are anion-specific porins, the binding site of which has a higher affinity for phosphate than chloride ions. Porin O has a higher affinity for polyphosphates, while porin P has a higher affinity for orthophosphate. In Pseudomonas aeruginosa, porin O was found to be expressed only under phosphate-starvation conditions during the stationary growth phase.
The public domain consists of all the creative works to which no exclusive intellectual property rights apply. Those rights may have expired, been forfeited, expressly waived, or may be inapplicable.

Pfam is a database of protein families that includes their annotations and multiple sequence alignments generated using hidden Markov models. The most recent version, Pfam 32.0, was released in September 2018 and contains 17,929 families.
InterPro is a database of protein families, domains and functional sites in which identifiable features found in known proteins can be applied to new protein sequences in order to functionally characterise them.