
A transmembrane protein is a type of integral membrane protein that spans the entirety of the cell membrane. Many transmembrane proteins function as gateways to permit the transport of specific substances across the membrane. They frequently undergo significant conformational changes to move a substance through the membrane. They are usually highly hydrophobic and aggregate and precipitate in water. They require detergents or nonpolar solvents for extraction, although some of them (beta-barrels) can be also extracted using denaturing agents.
A membrane transport protein is a membrane protein involved in the movement of ions, small molecules, and macromolecules, such as another protein, across a biological membrane. Transport proteins are integral transmembrane proteins; that is they exist permanently within and span the membrane across which they transport substances. The proteins may assist in the movement of substances by facilitated diffusion, active transport, osmosis, or reverse diffusion. The two main types of proteins involved in such transport are broadly categorized as either channels or carriers. Examples of channel/carrier proteins include the GLUT 1 uniporter, sodium channels, and potassium channels. The solute carriers and atypical SLCs are secondary active or facilitative transporters in humans. Collectively membrane transporters and channels are known as the transportome. Transportomes govern cellular influx and efflux of not only ions and nutrients but drugs as well.

Porins are beta barrel proteins that cross a cellular membrane and act as a pore, through which molecules can diffuse. Unlike other membrane transport proteins, porins are large enough to allow passive diffusion, i.e., they act as channels that are specific to different types of molecules. They are present in the outer membrane of gram-negative bacteria and some gram-positive mycobacteria, the outer membrane of mitochondria, and the outer chloroplast membrane.

The bacterial outer membrane is found in gram-negative bacteria. Gram-negative bacteria form two lipid bilayers in their cell envelopes - an inner membrane (IM) that encapsulates the cytoplasm, and an outer membrane (OM) that encapsulates the periplasm.
The Transporter Classification Database is an International Union of Biochemistry and Molecular Biology (IUBMB)-approved classification system for membrane transport proteins, including ion channels.

In protein structures, a beta barrel(β barrel) is a beta sheet composed of tandem repeats that twists and coils to form a closed toroidal structure in which the first strand is bonded to the last strand. Beta-strands in many beta-barrels are arranged in an antiparallel fashion. Beta barrel structures are named for resemblance to the barrels used to contain liquids. Most of them are water-soluble outer membrane proteins and frequently bind hydrophobic ligands in the barrel center, as in lipocalins. Others span cell membranes and are commonly found in porins. Porin-like barrel structures are encoded by as many as 2–3% of the genes in Gram-negative bacteria. It has been shown that more than 600 proteins with various function such as oxidase, dismutase, and amylase contain the beta barrel structure.
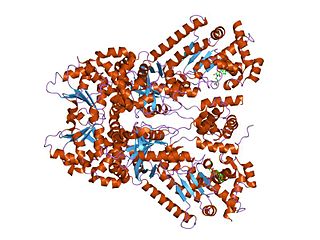
Anthrax toxin is a three-protein exotoxin secreted by virulent strains of the bacterium, Bacillus anthracis—the causative agent of anthrax. The toxin was first discovered by Harry Smith in 1954. Anthrax toxin is composed of a cell-binding protein, known as protective antigen (PA), and two enzyme components, called edema factor (EF) and lethal factor (LF). These three protein components act together to impart their physiological effects. Assembled complexes containing the toxin components are endocytosed. In the endosome, the enzymatic components of the toxin translocate into the cytoplasm of a target cell. Once in the cytosol, the enzymatic components of the toxin disrupts various immune cell functions, namely cellular signaling and cell migration. The toxin may even induce cell lysis, as is observed for macrophage cells. Anthrax toxin allows the bacteria to evade the immune system, proliferate, and ultimately kill the host animal. Research on anthrax toxin also provides insight into the generation of macromolecular assemblies, and on protein translocation, pore formation, endocytosis, and other biochemical processes.
Cytolysin refers to the substance secreted by microorganisms, plants or animals that is specifically toxic to individual cells, in many cases causing their dissolution through lysis. Cytolysins that have a specific action for certain cells are named accordingly. For instance, the cytolysins responsible for the destruction of red blood cells, thereby liberating hemoglobins, are named hemolysins, and so on. Cytolysins may be involved in immunity as well as in venoms.

Mitochondrial membrane transport proteins, also known as mitochondrial carrier proteins, are proteins which exist in the membranes of mitochondria. They serve to transport molecules and other factors, such as ions, into or out of the organelles. Mitochondria contain both an inner and outer membrane, separated by the inter-membrane space, or inner boundary membrane. The outer membrane is porous, whereas the inner membrane restricts the movement of all molecules. The two membranes also vary in membrane potential and pH. These factors play a role in the function of mitochondrial membrane transport proteins. There are 53 discovered human mitochondrial membrane transporters, with many others that are known to still need discovered.

Voltage-dependent anion channels, or mitochondrial porins, are a class of porin ion channel located on the outer mitochondrial membrane. There is debate as to whether or not this channel is expressed in the cell surface membrane.
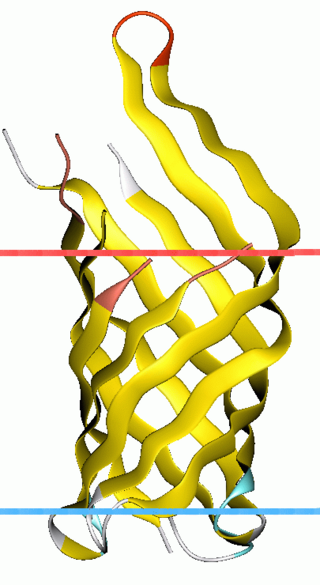
OmpA-like transmembrane domain is an evolutionarily conserved domain of bacterial outer membrane proteins. This domain consists of an eight-stranded beta barrel. OmpA is the predominant cell surface antigen in enterobacteria found in about 100,000 copies per cell. The expression of OmpA is tightly regulated by a variety of mechanisms. One mechanism by which OmpA expression is regulated in Vibrio species is by an antisense non-coding RNA called VrrA.

Maltoporins are bacterial outer membrane proteins of the porin family. Maltoporin forms a trimeric structure which facilitates the diffusion of maltodextrins across the outer membrane of Gram-negative bacteria. The membrane channel is formed by an antiparallel beta-barrel.

Virulence-related outer membrane proteins, or outer surface proteins (Osp) in some contexts, are expressed in the outer membrane of gram-negative bacteria and are essential to bacterial survival within macrophages and for eukaryotic cell invasion.
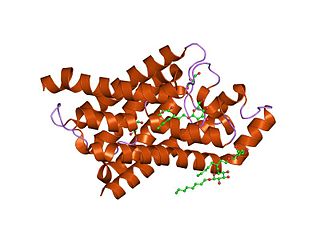
Major intrinsic proteins comprise a large superfamily of transmembrane protein channels that are grouped together on the basis of homology. The MIP superfamily includes three subfamilies: aquaporins, aquaglyceroporins and S-aquaporins.
- The aquaporins (AQPs) are water selective.
- The aquaglyceroporins are permeable to water, but also to other small uncharged molecules such as glycerol.
- The third subfamily, with little conserved amino acid sequences around the NPA boxes, include 'superaquaporins' (S-aquaporins).

Voltage-dependent anion-selective channel 1 (VDAC-1) is a beta barrel protein that in humans is encoded by the VDAC1 gene located on chromosome 5. It forms an ion channel in the outer mitochondrial membrane (OMM) and also the outer cell membrane. In the OMM, it allows ATP to diffuse out of the mitochondria into the cytoplasm. In the cell membrane, it is involved in volume regulation. Within all eukaryotic cells, mitochondria are responsible for synthesis of ATP among other metabolite needed for cell survival. VDAC1 therefore allows for communication between the mitochondrion and the cell mediating the balance between cell metabolism and cell death. Besides metabolic permeation, VDAC1 also acts as a scaffold for proteins such as hexokinase that can in turn regulate metabolism.
Omptins are a family of bacterial proteases. They are aspartate proteases, which cleave peptides with the use of a water molecule. Found in the outer membrane of gram-negative enterobacteria such as Shigella flexneri, Yersinia pestis, Escherichia coli, and Salmonella enterica. Omptins consist of a widely conserved beta barrel spanning the membrane with 5 extracellular loops. These loops are responsible for the various substrate specificities. These proteases rely upon binding of lipopolysaccharide for activity.
The thiol-activated Cholesterol-dependent Cytolysin(CDC) family is a member of the MACPF superfamily. Cholesterol dependent cytolysins are a family of β-barrel pore-forming exotoxins that are secreted by gram-positive bacteria. CDCs are secreted as water-soluble monomers of 50-70 kDa, that when bound to the target cell, form a circular homo-oligomeric complex containing as many as 40 monomers. Through multiple conformational changes, the β-barrel transmembrane structure is formed and inserted into the target cell membrane. The presence of cholesterol in the target membrane is required for pore formation, though the presence of cholesterol is not required by all CDCs for binding. For example, intermedilysin secreted by Streptococcus intermedius will bind only to target membranes containing a specific protein receptor, independent of the presence of cholesterol, but cholesterol is required by intermedilysin for pore formation. While the lipid environment of cholesterol in the membrane can affect toxin binding, the exact molecular mechanism that cholesterol regulates the cytolytic activity of the CDC is not fully understood.
Many bacteria secrete small iron-binding molecules called siderophores, which bind strongly to ferric ions. FepA is an integral bacterial outer membrane porin protein that belongs to outer membrane receptor family and provides the active transport of iron bound by the siderophore enterobactin from the extracellular space, into the periplasm of Gram-negative bacteria. FepA has also been shown to transport vitamin B12, and colicins B and D as well. This protein belongs to family of ligand-gated protein channels.

In molecular biology, the OmpA domain is a conserved protein domain with a beta/alpha/beta/alpha-beta(2) structure found in the C-terminal region of many Gram-negative bacterial outer membrane proteins, such as porin-like integral membrane proteins, small lipid-anchored proteins, and MotB proton channels. The N-terminal half of these proteins is variable although some of the proteins in this group have the OmpA-like transmembrane domain at the N terminus. OmpA from Escherichia coli is required for pathogenesis, and can interact with host receptor molecules. MotB serve two functions in E. coli, the MotA(4)-MotB(2) complex attaches to the cell wall via MotB to form the stator of the flagellar motor, and the MotA-MotB complex couples the flow of ions across the cell membrane to movement of the rotor.

OmpT is an aspartyl protease found on the outer membrane of Escherichia coli. OmpT is a subtype of the family of omptin proteases, which are found on some gram-negative species of bacteria.














