
A cell wall is a structural layer that surrounds some cell types, found immediately outside the cell membrane. It can be tough, flexible, and sometimes rigid. Primarily, it provides the cell with structural support, shape, protection, and functions as a selective barrier. Another vital role of the cell wall is to help the cell withstand osmotic pressure and mechanical stress. While absent in many eukaryotes, including animals, cell walls are prevalent in other organisms such as fungi, algae and plants, and are commonly found in most prokaryotes, with the exception of mollicute bacteria.

Gram stain, is a method of staining used to classify bacterial species into two large groups: gram-positive bacteria and gram-negative bacteria. It may also be used to diagnose a fungal infection. The name comes from the Danish bacteriologist Hans Christian Gram, who developed the technique in 1884.

In bacteriology, gram-positive bacteria are bacteria that give a positive result in the Gram stain test, which is traditionally used to quickly classify bacteria into two broad categories according to their type of cell wall.

Gram-negative bacteria are bacteria that unlike gram-positive bacteria do not retain the crystal violet stain used in the Gram staining method of bacterial differentiation. Their defining characteristic is their cell envelope, which consists of a thin peptidoglycan cell wall sandwiched between an inner (cytoplasmic) membrane and an outer membrane. These bacteria are found in all environments that support life on Earth.
Peptidoglycan or murein is a unique large macromolecule, a polysaccharide, consisting of sugars and amino acids that forms a mesh-like peptidoglycan layer (sacculus) that surrounds the bacterial cytoplasmic membrane. The sugar component consists of alternating residues of β-(1,4) linked N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM). Attached to the N-acetylmuramic acid is an oligopeptide chain made of three to five amino acids. The peptide chain can be cross-linked to the peptide chain of another strand forming the 3D mesh-like layer. Peptidoglycan serves a structural role in the bacterial cell wall, giving structural strength, as well as counteracting the osmotic pressure of the cytoplasm. This repetitive linking results in a dense peptidoglycan layer which is critical for maintaining cell form and withstanding high osmotic pressures, and it is regularly replaced by peptidoglycan production. Peptidoglycan hydrolysis and synthesis are two processes that must occur in order for cells to grow and multiply, a technique carried out in three stages: clipping of current material, insertion of new material, and re-crosslinking of existing material to new material.

Teichoic acids are bacterial copolymers of glycerol phosphate or ribitol phosphate and carbohydrates linked via phosphodiester bonds.
The periplasm is a concentrated gel-like matrix in the space between the inner cytoplasmic membrane and the bacterial outer membrane called the periplasmic space in gram-negative bacteria. Using cryo-electron microscopy it has been found that a much smaller periplasmic space is also present in gram-positive bacteria, between cell wall and the plasma membrane. The periplasm may constitute up to 40% of the total cell volume of gram-negative bacteria, but is a much smaller percentage in gram-positive bacteria.
An S-layer is a part of the cell envelope found in almost all archaea, as well as in many types of bacteria. The S-layers of both archaea and bacteria consists of a monomolecular layer composed of only one identical proteins or glycoproteins. This structure is built via self-assembly and encloses the whole cell surface. Thus, the S-layer protein can represent up to 15% of the whole protein content of a cell. S-layer proteins are poorly conserved or not conserved at all, and can differ markedly even between related species. Depending on species, the S-layers have a thickness between 5 and 25 nm and possess identical pores 2–8 nm in diameter.
Braun's lipoprotein, found in some gram-negative cell walls, is one of the most abundant membrane proteins; its molecular weight is about 7.2 kDa. It is bound at its C-terminal end by a covalent bond to the peptidoglycan layer and is embedded in the outer membrane by its hydrophobic head. BLP tightly links the two layers and provides structural integrity to the outer membrane.

Porins are beta barrel proteins that cross a cellular membrane and act as a pore, through which molecules can diffuse. Unlike other membrane transport proteins, porins are large enough to allow passive diffusion, i.e., they act as channels that are specific to different types of molecules. They are present in the outer membrane of gram-negative bacteria and some gram-positive mycobacteria, the outer membrane of mitochondria, and the outer chloroplast membrane.

The bacterial capsule is a large structure common to many bacteria. It is a polysaccharide layer that lies outside the cell envelope, and is thus deemed part of the outer envelope of a bacterial cell. It is a well-organized layer, not easily washed off, and it can be the cause of various diseases.
Pathogen-associated molecular patterns (PAMPs) are small molecular motifs conserved within a class of microbes, but not present in the host. They are recognized by toll-like receptors (TLRs) and other pattern recognition receptors (PRRs) in both plants and animals. This allows the innate immune system to recognize pathogens and thus, protect the host from infection.

The bacterial outer membrane is found in gram-negative bacteria. Gram-negative bacteria form two lipid bilayers in their cell envelopes - an inner membrane (IM) that encapsulates the cytoplasm, and an outer membrane (OM) that encapsulates the periplasm.
A bacterium, despite its simplicity, contains a well-developed cell structure which is responsible for some of its unique biological structures and pathogenicity. Many structural features are unique to bacteria and are not found among archaea or eukaryotes. Because of the simplicity of bacteria relative to larger organisms and the ease with which they can be manipulated experimentally, the cell structure of bacteria has been well studied, revealing many biochemical principles that have been subsequently applied to other organisms.
The cells of eukaryotic organisms are elaborately subdivided into functionally-distinct membrane-bound compartments. Some major constituents of eukaryotic cells are: extracellular space, plasma membrane, cytoplasm, nucleus, mitochondria, Golgi apparatus, endoplasmic reticulum (ER), peroxisome, vacuoles, cytoskeleton, nucleoplasm, nucleolus, nuclear matrix and ribosomes.
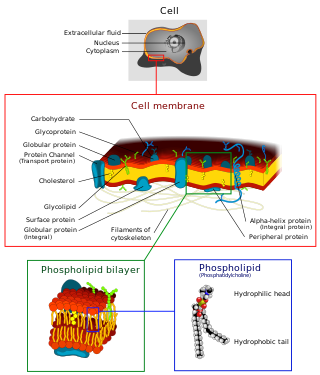
The cell membrane is a biological membrane that separates and protects the interior of a cell from the outside environment. The cell membrane consists of a lipid bilayer, made up of two layers of phospholipids with cholesterols interspersed between them, maintaining appropriate membrane fluidity at various temperatures. The membrane also contains membrane proteins, including integral proteins that span the membrane and serve as membrane transporters, and peripheral proteins that loosely attach to the outer (peripheral) side of the cell membrane, acting as enzymes to facilitate interaction with the cell's environment. Glycolipids embedded in the outer lipid layer serve a similar purpose. The cell membrane controls the movement of substances in and out of a cell, being selectively permeable to ions and organic molecules. In addition, cell membranes are involved in a variety of cellular processes such as cell adhesion, ion conductivity, and cell signalling and serve as the attachment surface for several extracellular structures, including the cell wall and the carbohydrate layer called the glycocalyx, as well as the intracellular network of protein fibers called the cytoskeleton. In the field of synthetic biology, cell membranes can be artificially reassembled.

Outer membrane vesicles (OMVs) are vesicles released from the outer membranes of Gram-negative bacteria. While Gram-positive bacteria release vesicles as well those vesicles fall under the broader category of bacterial membrane vesicles (MVs). OMVs were the first MVs to be discovered, and are distinguished from outer inner membrane vesicles (OIMVS), which are gram-negative baterial vesicles containing portions of both the outer and inner bacterial membrane. Outer membrane vesicles were first discovered and characterized using transmission-electron microscopy by Indian Scientist Prof. Smriti Narayan Chatterjee and J. Das in 1966-67. OMVs are ascribed the functionality to provide a manner to communicate among themselves, with other microorganisms in their environment and with the host. These vesicles are involved in trafficking bacterial cell signaling biochemicals, which may include DNA, RNA, proteins, endotoxins and allied virulence molecules. This communication happens in microbial cultures in oceans, inside animals, plants and even inside the human body.
The multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) flippase superfamily is a group of integral membrane protein families. The MOP flippase superfamily includes twelve distantly related families, six for which functional data are available:
- One ubiquitous family (MATE) specific for drugs - (TC# 2.A.66.1) The Multi Antimicrobial Extrusion (MATE) Family
- One (PST) specific for polysaccharides and/or their lipid-linked precursors in prokaryotes - (TC# 2.A.66.2) The Polysaccharide Transport (PST) Family
- One (OLF) specific for lipid-linked oligosaccharide precursors of glycoproteins in eukaryotes - (TC# 2.A.66.3) The Oligosaccharidyl-lipid Flippase (OLF) Family
- One (MVF) lipid-peptidoglycan precursor flippase involved in cell wall biosynthesis - (TC# 2.A.66.4) The Mouse Virulence Factor (MVF) Family
- One (AgnG) which includes a single functionally characterized member that extrudes the antibiotic, Agrocin 84 - (TC# 2.A.66.5) The Agrocin 84 Antibiotic Exporter (AgnG) Family
- And finally, one (Ank) that shuttles inorganic pyrophosphate (PPi) - (TC# 2.A.66.9) The Progressive Ankylosis (Ank) Family

Darobactin is an experimental antibiotic compound that may be effective against Gram-negative bacteria. If it can be developed into a human-compatible form it would be the first to come from an animal microbiome.
Undecaprenyl phosphate (UP), also known lipid-P, bactoprenol and C55-P., is a molecule with the primary function of trafficking polysaccharides across the cell membrane, largely contributing to the overall structure of the cell wall in Gram-positive bacteria. In some situations, UP can also be utilized to carry other cell-wall polysaccharides, but UP is the designated lipid carrier for peptidoglycan. During the process of carrying the peptidoglycan across the cell membrane, N-acetylglucosamine and N-acetylmuramic acid are linked to UP on the cytoplasmic side of the membrane before being carried across. UP works in a cycle of phosphorylation and dephosphorylation as the lipid carrier gets used, recycled, and reacts with undecaprenyl phosphate. This type of synthesis is referred to as de novo synthesis where a complex molecule is created from simpler molecules as opposed to a complete recycle of the entire structure.












