Peptidoglycan or murein is a unique large macromolecule, a polysaccharide, consisting of sugars and amino acids that forms a mesh-like peptidoglycan layer outside the plasma membrane, the rigid cell wall characteristic of most bacteria. The sugar component consists of alternating residues of β-(1,4) linked N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM). Attached to the N-acetylmuramic acid is an oligopeptide chain made of three to five amino acids. The peptide chain can be cross-linked to the peptide chain of another strand forming the 3D mesh-like layer. Peptidoglycan serves a structural role in the bacterial cell wall, giving structural strength, as well as counteracting the osmotic pressure of the cytoplasm. This repetitive linking results in a dense peptidoglycan layer which is critical for maintaining cell form and withstanding high osmotic pressures, and it is regularly replaced by peptidoglycan production. Peptidoglycan hydrolysis and synthesis are two processes that must occur in order for cells to grow and multiply, a technique carried out in three stages: clipping of current material, insertion of new material, and re-crosslinking of existing material to new material.

Teichoic acids are bacterial copolymers of glycerol phosphate or ribitol phosphate and carbohydrates linked via phosphodiester bonds.

N-Acetylglucosamine (GlcNAc) is an amide derivative of the monosaccharide glucose. It is a secondary amide between glucosamine and acetic acid. It is significant in several biological systems.

Pseudopeptidoglycan is a major cell wall component of some Archaea that differs from bacterial peptidoglycan in chemical structure, but resembles bacterial peptidoglycan in function and physical structure. Pseudopeptidoglycan, in general, is only present in a few methanogenic archaea. The basic components are N-acetylglucosamine and N-acetyltalosaminuronic acid, which are linked by β-1,3-glycosidic bonds.

Lysins, also known as endolysins or murein hydrolases, are hydrolytic enzymes produced by bacteriophages in order to cleave the host's cell wall during the final stage of the lytic cycle. Lysins are highly evolved enzymes that are able to target one of the five bonds in peptidoglycan (murein), the main component of bacterial cell walls, which allows the release of progeny virions from the lysed cell. Cell-wall-containing Archaea are also lysed by specialized pseudomurein-cleaving lysins, while most archaeal viruses employ alternative mechanisms. Similarly, not all bacteriophages synthesize lysins: some small single-stranded DNA and RNA phages produce membrane proteins that activate the host's autolytic mechanisms such as autolysins.

Fosfomycin, sold under the brand name Monurol among others, is an antibiotic primarily used to treat lower urinary tract infections. It is not indicated for kidney infections. Occasionally it is used for prostate infections. It is generally taken by mouth.
In enzymology, a UDP-N-acetylmuramate—L-alanine ligase is an enzyme that catalyzes the chemical reaction
In enzymology, a UDP-N-acetylmuramoyl-L-alanine—D-glutamate ligase is an enzyme that catalyzes the chemical reaction
In enzymology, a UDP-N-acetylmuramoyl-L-alanyl-D-glutamate—L-lysine ligase is an enzyme that catalyzes the chemical reaction
In enzymology, a UDP-N-acetylmuramoyl-tripeptide—D-alanyl-D-alanine ligase is an enzyme that catalyzes the chemical reaction

In enzymology, N-acetylglucosamine-6-phosphate deacetylase (EC 3.5.1.25), also known as GlcNAc-6-phosphate deacetylase or NagA, is an enzyme that catalyzes the deacetylation of N-acetylglucosamine-6-phosphate (GlcNAc-6-P) to glucosamine-6-phosphate (GlcN-6-P):
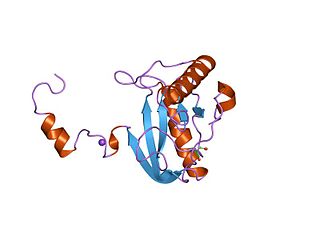
In enzymology, a N-acetylmuramoyl-L-alanine amidase is an enzyme that catalyzes a chemical reaction that cleaves the link between N-acetylmuramoyl residues and L-amino acid residues in certain cell-wall glycopeptides.

In enzymology, an UDP-N-acetylglucosamine 1-carboxyvinyltransferase is an enzyme that catalyzes the first committed step in peptidoglycan biosynthesis of bacteria:

Peptidoglycan recognition protein 2(PGLYRP2) is an enzyme, N-acetylmuramoyl-L-alanine amidase (NAMLAA), that hydrolyzes bacterial cell wall peptidoglycan and is encoded by the PGLYRP2 gene.
The bacterial cell wall provides strength and rigidity to counteract internal osmotic pressure, and protection against the environment. The peptidoglycan layer gives the cell wall its strength, and helps maintain the overall shape of the cell. The basic peptidoglycan structure of both Gram-positive and Gram-negative bacteria comprises a sheet of glycan chains connected by short cross-linking polypeptides. Biosynthesis of peptidoglycan is a multi-step process comprising three main stages:
- formation of UDP-N-acetylmuramic acid (UDPMurNAc) from N-acetylglucosamine (GlcNAc).
- addition of a short polypeptide chain to the UDPMurNAc.
- addition of a second GlcNAc to the disaccharide-pentapeptide building block and transport of this unit through the cytoplasmic membrane and incorporation into the growing peptidoglycan layer.
N-acetylmuramoyl-L-alanyl-D-glutamyl-L-lysyl-(N6-triglycine)-D-alanyl-D-alanine-diphosphoundecaprenyl-N-acetylglucosamine:glycine glycyltransferase (EC 2.3.2.18, femB (gene)) is an enzyme with systematic name N-acetylmuramoyl-L-alanyl-D-glutamyl-L-lysyl-(N6-triglycine)-D-alanyl-D-alanine-ditrans,octacis-diphosphoundecaprenyl-N-acetylglucosamine:glycine glycyltransferase. This enzyme catalyses the following chemical reaction
N-acetylmuramic acid 6-phosphate etherase (EC 4.2.1.126, MurNAc-6-P etherase, MurQ) is an enzyme with systematic name (R)-lactate hydro-lyase (adding N-acetyl-D-glucosamine 6-phosphate; N-acetylmuramate 6-phosphate-forming). This enzyme catalyses the following chemical reaction

Lipid II is a precursor molecule in the synthesis of the cell wall of bacteria. It is a peptidoglycan, which is amphipathic and named for its bactoprenol hydrocarbon chain, which acts as a lipid anchor, embedding itself in the bacterial cell membrane. Lipid II must translocate across the cell membrane to deliver and incorporate its disaccharide-pentapeptide "building block" into the peptidoglycan mesh. Lipid II is the target of several antibiotics.

Peptidoglycan recognition protein 3 is an antibacterial and anti-inflammatory innate immunity protein that in humans is encoded by the PGLYRP3 gene.
Undecaprenyl phosphate (UP), also known lipid-P, bactoprenol and C55-P., is a molecule with the primary function of trafficking polysaccharides across the cell membrane, largely contributing to the overall structure of the cell wall in Gram-positive bacteria. In some situations, UP can also be utilized to carry other cell-wall polysaccharides, but UP is the designated lipid carrier for peptidoglycan. During the process of carrying the peptidoglycan across the cell membrane, N-acetylglucosamine and N-acetylmuramic acid are linked to UP on the cytoplasmic side of the membrane before being carried across. UP works in a cycle of phosphorylation and dephosphorylation as the lipid carrier gets used, recycled, and reacts with undecaprenyl phosphate. This type of synthesis is referred to as de novo synthesis where a complex molecule is created from simpler molecules as opposed to a complete recycle of the entire structure.











