| Gracilicutes | |
|---|---|
 | |
| Escherichia coli cells magnified 25,000 times | |
| Scientific classification | |
| Domain: | Bacteria |
| (unranked): | Gracilicutes Gibbons and Murray 1978 [1] |
| Superphyla/Phyla | |
Various definitions, see text | |
Gracilicutes (Latin: gracilis, slender, and cutis, skin, referring to the cell wall) is a clade in bacterial phylogeny. [2]
Traditionally gram staining results were most commonly used as a classification tool, consequently until the advent of molecular phylogeny, the Kingdom Monera (as the domains Bacteria and Archaea were known then) was divided into four phyla, [1] [3]
- Gracilicutes (gram-negative, it is split in many groups, but some authors still use it in a narrower sense)
- Firmacutes [sic] (gram-positive, subsequently corrected to Firmicutes, [4] today it excludes the Actinomycetota)
- Mollicutes (gram variable, later renamed Tenericutes and now Mycoplasmatota, e.g. Mycoplasma )
- Mendosicutes (uneven gram stain, "methanogenic bacteria" now known as methanogens and classed as Archaea)
This classification system was abandoned in favour of the three-domain system based on molecular phylogeny started by C. Woese. [5] [6]
Using hand-drawn schematics rather than standard molecular phylogenetic analysis, Gracilicutes was revived in 2006 by Cavalier-Smith as an infrakindgom containing the phyla Spirochaetota, Sphingobacteria (FCB), Planctobacteria (PVC), and Proteobacteria. [7] It is a gram-negative clade that branched off from other bacteria just before the evolutionary loss of the outer membrane or capsule, and just after the evolution of flagella. [7] Most notably, this author assumed an unconventional tree of life placing Chloroflexota near the origin of life and Archaea as a close relative of Actinomycetota. This taxon is not generally accepted and the three-domain system is followed. [8]
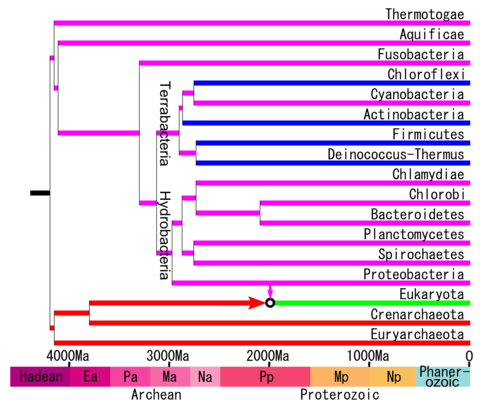
A taxon called Hydrobacteria was defined in 2009 from a molecular phylogenetic analysis of core genes. It is in contrast to the other major group of eubacteria called Terrabacteria . [9] Some researchers have used the name Gracilicutes in place of Hydrobacteria , but this does not agree with the original description of Gracilicutes by Gibbons and Murray, noted above, which included cyanobacteria and did not follow the three-domain system. Also as noted above, the use of Gracilicutes by Cavalier-Smith can be rejected because it was a major alteration of an earlier taxonomic name, was not based on a statistical analysis, and did not follow the three-domain system. The most recent genomic analyses have supported the division of Bacteria into two major superphyla, corresponding to Terrabacteria and Hydrobacteria . [10] [11]