Related Research Articles

Health informatics is the field of science and engineering that aims at developing methods and technologies for the acquisition, processing, and study of patient data, which can come from different sources and modalities, such as electronic health records, diagnostic test results, medical scans. The health domain provides an extremely wide variety of problems that can be tackled using computational techniques.
An electronic health record (EHR) is the systematized collection of patient and population electronically stored health information in a digital format. These records can be shared across different health care settings. Records are shared through network-connected, enterprise-wide information systems or other information networks and exchanges. EHRs may include a range of data, including demographics, medical history, medication and allergies, immunization status, laboratory test results, radiology images, vital signs, personal statistics like age and weight, and billing information.
A clinical terminology server is a terminology server, which contains and provides access to clinical terminology.
The Unified Medical Language System (UMLS) is a compendium of many controlled vocabularies in the biomedical sciences. It provides a mapping structure among these vocabularies and thus allows one to translate among the various terminology systems; it may also be viewed as a comprehensive thesaurus and ontology of biomedical concepts. UMLS further provides facilities for natural language processing. It is intended to be used mainly by developers of systems in medical informatics.
Biomedical text mining refers to the methods and study of how text mining may be applied to texts and literature of the biomedical and molecular biology domains. As a field of research, biomedical text mining incorporates ideas from natural language processing, bioinformatics, medical informatics and computational linguistics. The strategies developed through studies in this field are frequently applied to the biomedical and molecular biology literature available through services such as PubMed.

SNOMED CT or SNOMED Clinical Terms is a systematically organized computer-processable collection of medical terms providing codes, terms, synonyms and definitions used in clinical documentation and reporting. SNOMED CT is considered to be the most comprehensive, multilingual clinical healthcare terminology in the world. The primary purpose of SNOMED CT is to encode the meanings that are used in health information and to support the effective clinical recording of data with the aim of improving patient care. SNOMED CT provides the core general terminology for electronic health records. SNOMED CT comprehensive coverage includes: clinical findings, symptoms, diagnoses, procedures, body structures, organisms and other etiologies, substances, pharmaceuticals, devices and specimens.
Classification Commune des Actes Médicaux is a French medical classification for clinical procedures. Starting in 2005, the CCAM serves as the reimbursement classification for clinicians. The CCAM was evaluated using OpenGALEN tools and technologies.
Public health informatics has been defined as the systematic application of information and computer science and technology to public health practice, research, and learning. It is one of the subdomains of health informatics.

Carole Anne Goble, is a British academic who is Professor of Computer Science at the University of Manchester. She is principal investigator (PI) of the myGrid, BioCatalogue and myExperiment projects and co-leads the Information Management Group (IMG) with Norman Paton.

Alan L. Rector is a Professor of Medical Informatics in the Department of Computer Science at the University of Manchester in the UK.
MEDCIN, a system of standardized medical terminology, is a proprietary medical vocabulary and was developed by Medicomp Systems, Inc. MEDCIN is a point-of-care terminology, intended for use in Electronic Health Record (EHR) systems, and it includes over 280,000 clinical data elements encompassing symptoms, history, physical examination, tests, diagnoses and therapy. This clinical vocabulary contains over 38 years of research and development as well as the capability to cross map to leading codification systems such as SNOMED CT, CPT, ICD-9-CM/ICD-10-CM, DSM, LOINC, CDT, CVX, and the Clinical Care Classification (CCC) System for nursing and allied health.

OpenMRS is a collaborative open-source project to develop software to support the delivery of health care in developing countries.
Health information technology (HIT) is health technology, particularly information technology, applied to health and health care. It supports health information management across computerized systems and the secure exchange of health information between consumers, providers, payers, and quality monitors. Based on a 2008 report on a small series of studies conducted at four sites that provide ambulatory care – three U.S. medical centers and one in the Netherlands, the use of electronic health records (EHRs) was viewed as the most promising tool for improving the overall quality, safety and efficiency of the health delivery system.
The Omaha System is a standardized health care terminology consisting of an assessment component, a care plan/services component, and an evaluation component. Approximately 22,000 health care practitioners, educators, and researchers use Omaha System to improve clinical practice, structure documentation, and analyze secondary data. Omaha System users from Canada, China, The Czech Republic, Estonia, Hong Kong, Japan, Mexico, New Zealand, The Netherlands, Turkey, the United States, and Wales, have presented at Omaha System International Conferences.
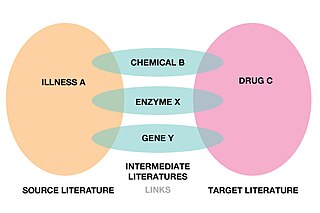
Literature-based discovery (LBD), also called literature-related discovery (LRD) is a form of knowledge extraction and automated hypothesis generation that uses papers and other academic publications to find new relationships between existing knowledge. Literature-based discovery aims to discover new knowledge by connecting information which have been explicitly stated in literature to deduce connections which have not been explicitly stated.
The Clinical Care Classification (CCC) System is a standardized, coded nursing terminology that identifies the discrete elements of nursing practice. The CCC provides a unique framework and coding structure. Used for documenting the plan of care; following the nursing process in all health care settings.
Translational bioinformatics (TBI) is an emerging field in the study of health informatics, focused on the convergence of molecular bioinformatics, biostatistics, statistical genetics and clinical informatics. Its focus is on applying informatics methodology to the increasing amount of biomedical and genomic data to formulate knowledge and medical tools, which can be utilized by scientists, clinicians, and patients. Furthermore, it involves applying biomedical research to improve human health through the use of computer-based information system. TBI employs data mining and analyzing biomedical informatics in order to generate clinical knowledge for application. Clinical knowledge includes finding similarities in patient populations, interpreting biological information to suggest therapy treatments and predict health outcomes.

Dipak Kalra is President of the European Institute for Health Records and of the European Institute for Innovation through Health Data. He undertakes international research and standards development, and advises on adoption strategies, relating to Electronic Health Records.
The inability of an ontology to include or encode every concept of interest to users of an ontology is known as the "content completeness problem". The problem stems from the greater expressiveness of natural language relative to the finite enumeration of concepts present in an ontology. A specific instance of this problem may be temporarily resolved by adding missing concept/s to the target ontology, but this approach is not a general solution because in practice:
- There is a lag time between the appearance and usage of a concept in a domain and its incorporation into an ontology.
- In large ontologies it is difficult to know if the concept is already present

Dean Forrest Sittig is an American biomedical informatician specializing in clinical informatics. He is a professor in Biomedical Informatics at the University of Texas Health Science Center at Houston and Executive Director of the Clinical Informatics Research Collaborative (CIRCLE). Sittig was elected as a fellow of the American College of Medical Informatics in 1992, the Healthcare Information and Management Systems Society in 2011, and was a founding member of the International Academy of Health Sciences Informatics in 2017. Since 2004, he has worked with Joan S. Ash, a professor at Oregon Health & Science University to interview several Pioneers in Medical Informatics, including G. Octo Barnett, MD, Morris F. Collen, MD, Donald E. Detmer, MD, Donald A. B. Lindberg, MD, Nina W. Matheson, ML, DSc, Clement J. McDonald, MD, and Homer R. Warner, MD, PhD.
References
- Rector, A.; Rogers, J.; Zanstra, P.; Van Der Haring, E. (2003). "OpenGALEN: Open source medical terminology and tools". AMIA Annual Symposium Proceedings. 2003: 982. PMC 1480228 . PMID 14728486.
- Rector, A.; Solomon, W.; Nowlan, W.; Rush, T.; Zanstra, P.; Claassen, W. (1995). "A Terminology Server for medical language and medical information systems". Methods of Information in Medicine. 34 (1–2): 147–157. doi:10.1055/s-0038-1634569. PMID 9082124. S2CID 7978610.
- Rector, A.; Zanstra, P.; Solomon, W.; Rogers, J.; Baud, R.; Ceusters, W.; Claassen, W.; Kirby, J.; Rodrigues, J.; Rossi Mori, A. R.; Van Der Haring, E. J.; Wagner, J. (1998). "Reconciling users' needs and formal requirements: Issues in developing a reusable ontology for medicine". IEEE Transactions on Information Technology in Biomedicine. 2 (4): 229–242. doi:10.1109/4233.737578. PMID 10719533. S2CID 10702025.
- Rogers, J.; Roberts, A.; Solomon, D.; Van Der Haring, E.; Wroe, C.; Zanstra, P.; Rector, A. (2001). "GALEN ten years on: Tasks and supporting tools". Studies in Health Technology and Informatics. 84 (Pt 1): 256–260. PMID 11604744.
- Ten Napel, H.; Rogers, J. (2001). "Assessment of the GALEN methodology on holistic classifications for professions allied to medicine". Studies in Health Technology and Informatics. 84 (Pt 2): 1369–1373. PMID 11604951.
- Rogers, J.; Rector, A. (2000). "GALEN's model of parts and wholes: Experience and comparisons". AMIA Annual Symposium Proceedings: 714–718. PMC 2243933 . PMID 11079977.
- Solomon, W.; Roberts, A.; Rogers, J.; Wroe, C.; Rector, A. (2000). "Having our cake and eating it too: How the GALEN Intermediate Representation reconciles internal complexity with users' requirements for appropriateness and simplicity". AMIA Annual Symposium Proceedings: 819–823. PMC 2244105 . PMID 11079998.
- Rogers, J. (2006). "Quality assurance of medical ontologies". Methods of Information in Medicine. 45 (3): 267–274. doi:10.1055/s-0038-1634078. PMID 16685334. S2CID 9398450.