
Sulfur-reducing bacteria are microorganisms able to reduce elemental sulfur (S0) to hydrogen sulfide (H2S). These microbes use inorganic sulfur compounds as electron acceptors to sustain several activities such as respiration, conserving energy and growth, in absence of oxygen. The final product of these processes, sulfide, has a considerable influence on the chemistry of the environment and, in addition, is used as electron donor for a large variety of microbial metabolisms. Several types of bacteria and many non-methanogenic archaea can reduce sulfur. Microbial sulfur reduction was already shown in early studies, which highlighted the first proof of S0 reduction in a vibrioid bacterium from mud, with sulfur as electron acceptor and H
2 as electron donor. The first pure cultured species of sulfur-reducing bacteria, Desulfuromonas acetoxidans, was discovered in 1976 and described by Pfennig Norbert and Biebel Hanno as an anaerobic sulfur-reducing and acetate-oxidizing bacterium, not able to reduce sulfate. Only few taxa are true sulfur-reducing bacteria, using sulfur reduction as the only or main catabolic reaction. Normally, they couple this reaction with the oxidation of acetate, succinate or other organic compounds. In general, sulfate-reducing bacteria are able to use both sulfate and elemental sulfur as electron acceptors. Thanks to its abundancy and thermodynamic stability, sulfate is the most studied electron acceptor for anaerobic respiration that involves sulfur compounds. Elemental sulfur, however, is very abundant and important, especially in deep-sea hydrothermal vents, hot springs and other extreme environments, making its isolation more difficult. Some bacteria – such as Proteus, Campylobacter, Pseudomonas and Salmonella – have the ability to reduce sulfur, but can also use oxygen and other terminal electron acceptors.
Fibrobacterota is a small bacterial phylum which includes many of the major rumen bacteria, allowing for the degradation of plant-based cellulose in ruminant animals. Members of this phylum were categorized in other phyla. The genus Fibrobacter was removed from the genus Bacteroides in 1988.
In biology, syntrophy, syntrophism, or cross-feeding is the cooperative interaction between at least two microbial species to degrade a single substrate. This type of biological interaction typically involves the transfer of one or more metabolic intermediates between two or more metabolically diverse microbial species living in close proximity to each other. Thus, syntrophy can be considered an obligatory interdependency and a mutualistic metabolism between different microbial species, wherein the growth of one partner depends on the nutrients, growth factors, or substrates provided by the other(s).
Paracoccus denitrificans, is a coccoid bacterium known for its nitrate reducing properties, its ability to replicate under conditions of hypergravity and for being a relative of the eukaryotic mitochondrion.
Oxalobacter formigenes is a Gram negative oxalate-degrading anaerobic bacterium that was first isolated from the gastrointestinal tract of a sheep in 1985. To date, the bacterium has been found to colonize the large intestines of numerous vertebrates, including humans, and has even been isolated from freshwater sediment. It processes oxalate by decarboxylation into formate, producing energy for itself in the process.
In enzymology, a formyl-CoA transferase is an enzyme that catalyzes the chemical reaction
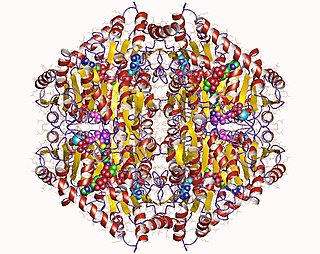
The enzyme oxalyl-CoA decarboxylase (OXC) (EC 4.1.1.8), primarily produced by the gastrointestinal bacterium Oxalobacter formigenes, catalyzes the chemical reaction
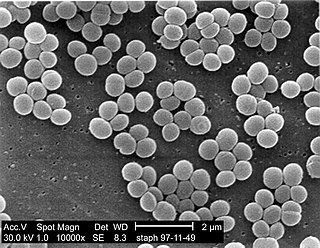
Staphylococcus is a genus of Gram-positive bacteria in the family Staphylococcaceae from the order Bacillales. Under the microscope, they appear spherical (cocci), and form in grape-like clusters. Staphylococcus species are facultative anaerobic organisms.
Oxalobacter is a genus of Gram-negative bacteria in the Oxalobacteraceae family. The species are chemoorganotrophs and strictly anaerobic. They are found in rumens of animals such as cattle and in feces of other animals, rodents, and humans. Oxalobacter species have also been isolated from marine sources, including from fresh water samples. These bacteria are characterized by their ability to metabolize oxalate.
Oxalobacter vibrioformis is an oxalate-degrading anaerobic bacterium that was isolated from anoxic freshwater sediments. O. vibrioformis is a Gram-negative, non-spore-forming, motile, vibrioid rod which belongs to the genus Oxalobacter. O. vibrioformis uses oxalate and oxamate as its sole source of energy and acetate as its main source of carbon.
Synergistes jonesii is a species of bacteria, the type species of its genus. It is a rumen bacterium that degrades toxic pyridinediols including mimosine. It is obligately anaerobic, gram-negative and rod-shaped. It was discovered in 1981 by Raymond J. Jones in Hawaii and Jones' hypothesis was proven in 1986 by himself and R. G. Megarrity.
Rhodoferax is a genus of Betaproteobacteria belonging to the purple nonsulfur bacteria. Originally, Rhodoferax species were included in the genus Rhodocyclus as the Rhodocyclus gelatinous-like group. The genus Rhodoferax was first proposed in 1991 to accommodate the taxonomic and phylogenetic discrepancies arising from its inclusion in the genus Rhodocyclus. Rhodoferax currently comprises four described species: R. fermentans, R. antarcticus, R. ferrireducens, and R. saidenbachensis. R. ferrireducens, lacks the typical phototrophic character common to two other Rhodoferax species. This difference has led researchers to propose the creation of a new genus, Albidoferax, to accommodate this divergent species. The genus name was later corrected to Albidiferax. Based on geno- and phenotypical characteristics, A. ferrireducens was reclassified in the genus Rhodoferax in 2014. R. saidenbachensis, a second non-phototrophic species of the genus Rhodoferax was described by Kaden et al. in 2014.
Oleispira antarctica is a hydrocarbonoclastic marine bacterium, the type species in its genus. It is psychrophilic, aerobic and Gram-negative, with polar flagellum. Its genome has been sequenced and from this information, it has been recognized as a potentially important organism capable of oil degradation in the deep sea.
Symbiobacterium thermophilum is a symbiotic thermophile that depends on co-culture with a Bacillus strain for growth. It is Gram-negative and tryptophanase-positive, with type strain T(T). It is the type species of its genus. Symbiobacterium is related to the Gram-positive Bacillota and Actinomycetota, but belongs to a lineage that is distinct from both.S. thermophilum has a bacillus shaped cell structure with no flagella. This bacterium is located throughout the environment in soils and fertilizers.
Oscillibacter valericigenes is a species of mesophilic bacterium identified in the alimentary canal of Japanese Corbicula clams. It is Gram-negative and anaerobic, with a straight to slightly curved rod-like morphology, and is motile with petritrichous flagella. It was not observed in culture to form spores.
Anaerococcus is a genus of bacteria. Its type species is Anaerococcus prevotii. These bacteria are Gram-positive and strictly anaerobic. The genus Anaerococcus was proposed in 2001. Its genome was sequenced in August 2009. The genus Anaerococcus is one of six genera classified within the group GPAC. These six genera are found in the human body as part of the commensal human microbiota.
Polaribacter is a genus in the family Flavobacteriaceae. They are gram-negative, aerobic bacteria that can be heterotrophic, psychrophilic or mesophilic. Most species are non-motile and species range from ovoid to rod-shaped. Polaribacter forms yellow- to orange-pigmented colonies. They have been mostly adapted to cool marine ecosystems, and their optimal growth range is at a temperature between 10 and 32 °C and at a pH of 7.0 to 8.0. They are oxidase and catalase-positive and are able to grow using carbohydrates, amino acids, and organic acids.
Cytophagales is an order of non-spore forming, rod-shaped, Gram-negative bacteria that move through a gliding or flexing motion. These chemoorganotrophs are important remineralizers of organic materials into micronutrients. They are widely dispersed in the environment, found in ecosystems including soil, freshwater, seawater and sea ice. Cytophagales is included in the Bacteroidota phylum.
Oxalobacter aliiformigenes is a Gram negative, non-spore-forming, oxalate-degrading anaerobic bacterium that was first isolated from human fecal samples. O. aliiformigenes consumes oxalate as its main carbon source but is negative for indole production and negative for sulfate and nitrate reduction. Cells appear rod shaped, though occasionally present as curved, and do not possess flagella.
Oxalobacter paraformigenes is a Gram negative, non-spore-forming, oxalate-degrading anaerobic bacterium that was first isolated from human fecal samples. O. paraformigenes may have a role in calcium oxalate kidney stone disease because of its unique ability to utilize oxalate as its primary carbon source.


