
MicroRNA (miRNA) are small, single-stranded, non-coding RNA molecules containing 21 to 23 nucleotides. Found in plants, animals and some viruses, miRNAs are involved in RNA silencing and post-transcriptional regulation of gene expression. miRNAs base-pair to complementary sequences in mRNA molecules, then gene silence said mRNA molecules by one or more of the following processes:
- Cleavage of mRNA strand into two pieces,
- Destabilization of mRNA by shortening its poly(A) tail, or
- Translation of mRNA into proteins.

Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, proteins or non-coding RNA, and ultimately affect a phenotype. These products are often proteins, but in non-protein-coding genes such as transfer RNA (tRNA) and small nuclear RNA (snRNA), the product is a functional non-coding RNA. Gene expression is summarized in the central dogma of molecular biology first formulated by Francis Crick in 1958, further developed in his 1970 article, and expanded by the subsequent discoveries of reverse transcription and RNA replication.

A non-coding RNA (ncRNA) is a functional RNA molecule that is not translated into a protein. The DNA sequence from which a functional non-coding RNA is transcribed is often called an RNA gene. Abundant and functionally important types of non-coding RNAs include transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs), as well as small RNAs such as microRNAs, siRNAs, piRNAs, snoRNAs, snRNAs, exRNAs, scaRNAs and the long ncRNAs such as Xist and HOTAIR.

The Argonaute protein family, first discovered for its evolutionarily conserved stem cell function, plays a central role in RNA silencing processes as essential components of the RNA-induced silencing complex (RISC). RISC is responsible for the gene silencing phenomenon known as RNA interference (RNAi). Argonaute proteins bind different classes of small non-coding RNAs, including microRNAs (miRNAs), small interfering RNAs (siRNAs) and Piwi-interacting RNAs (piRNAs). Small RNAs guide Argonaute proteins to their specific targets through sequence complementarity, which then leads to mRNA cleavage, translation inhibition, and/or the initiation of mRNA decay.
RNA silencing or RNA interference refers to a family of gene silencing effects by which gene expression is negatively regulated by non-coding RNAs such as microRNAs. RNA silencing may also be defined as sequence-specific regulation of gene expression triggered by double-stranded RNA (dsRNA). RNA silencing mechanisms are conserved among most eukaryotes. The most common and well-studied example is RNA interference (RNAi), in which endogenously expressed microRNA (miRNA) or exogenously derived small interfering RNA (siRNA) induces the degradation of complementary messenger RNA. Other classes of small RNA have been identified, including piwi-interacting RNA (piRNA) and its subspecies repeat associated small interfering RNA (rasiRNA).

MicroRNA (miRNA) precursor miR156 is a family of plant non-coding RNA. This microRNA has now been predicted or experimentally confirmed in a range of plant species. Animal miRNAs are transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give a ~22 nucleotide product. miR156 functions in the induction of flowering by suppressing the transcripts of SQUAMOSA-PROMOTER BINDING LIKE (SPL) transcription factors gene family. It was suggested that the loading into ARGONAUTE1 and ARGONAUTE5 is required for miR156 functionality in Arabidopsis thaliana. In plants the precursor sequences may be longer, and the carpel factory (caf) enzyme appears to be involved in processing. In this case the mature sequence comes from the 5' arm of the precursor, and both Arabidopsis thaliana and rice genomes contain a number of related miRNA precursors which give rise to almost identical mature sequences. The extents of the hairpin precursors are not generally known and are estimated based on hairpin prediction. The products are thought to have regulatory roles through complementarity to mRNA.

The mir-2 microRNA family includes the microRNA genes mir-2 and mir-13. Mir-2 is widespread in invertebrates, and it is the largest family of microRNAs in the model species Drosophila melanogaster. MicroRNAs from this family are produced from the 3' arm of the precursor hairpin. Leaman et al. showed that the miR-2 family regulates cell survival by translational repression of proapoptotic factors. Based on computational prediction of targets, a role in neural development and maintenance has been suggested.
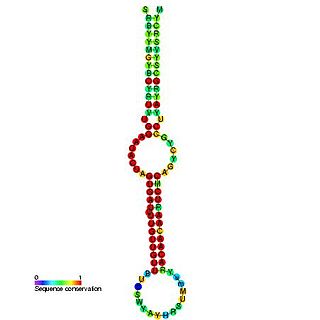
This family represents the microRNA (miRNA) precursor mir-7. This miRNA has been predicted or experimentally confirmed in a wide range of species. miRNAs are transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give a ~22 nucleotide product. In this case the mature sequence comes from the 5' arm of the precursor. The extents of the hairpin precursors are not generally known and are estimated based on hairpin prediction. The involvement of Dicer in miRNA processing suggests a relationship with the phenomenon of RNA interference.

MicrobesOnline is a publicly and freely accessible website that hosts multiple comparative genomic tools for comparing microbial species at the genomic, transcriptomic and functional levels. MicrobesOnline was developed by the Virtual Institute for Microbial Stress and Survival, which is based at the Lawrence Berkeley National Laboratory in Berkeley, California. The site was launched in 2005, with regular updates until 2011.
GeneCards is a database of human genes, which provides genomic, proteomic, transcriptomic, genetic, medical, and functional information on all known and predicted human genes. It is being developed and maintained by the Crown Human Genome Center at the Weizmann Institute of Science, in collaboration with LifeMap Sciences.
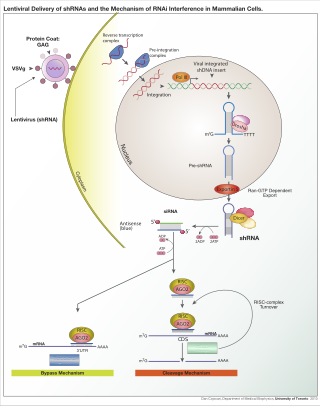
RNA interference (RNAi) is a biological process in which RNA molecules are involved in sequence-specific suppression of gene expression by double-stranded RNA, through translational or transcriptional repression. Historically, RNAi was known by other names, including co-suppression, post-transcriptional gene silencing (PTGS), and quelling. The detailed study of each of these seemingly different processes elucidated that the identity of these phenomena were all actually RNAi. Andrew Fire and Craig C. Mello shared the 2006 Nobel Prize in Physiology or Medicine for their work on RNAi in the nematode worm Caenorhabditis elegans, which they published in 1998. Since the discovery of RNAi and its regulatory potentials, it has become evident that RNAi has immense potential in suppression of desired genes. RNAi is now known as precise, efficient, stable and better than antisense therapy for gene suppression. Antisense RNA produced intracellularly by an expression vector may be developed and find utility as novel therapeutic agents.

In bioinformatics, miRBase is a biological database that acts as an archive of microRNA sequences and annotations. As of September 2010 it contained information about 15,172 microRNAs. This number has risen to 38,589 by March 2018. The miRBase registry provides a centralised system for assigning new names to microRNA genes.

In molecular biology, miR-137 is a short non-coding RNA molecule that functions to regulate the expression levels of other genes by various mechanisms. miR-137 is located on human chromosome 1p22 and has been implicated to act as a tumor suppressor in several cancer types including colorectal cancer, squamous cell carcinoma and melanoma via cell cycle control.
This microRNA database and microRNA targets databases is a compilation of databases and web portals and servers used for microRNAs and their targets. MicroRNAs (miRNAs) represent an important class of small non-coding RNAs (ncRNAs) that regulate gene expression by targeting messenger RNAs.
Degradome sequencing (Degradome-Seq), also referred to as parallel analysis of RNA ends (PARE), is a modified version of 5'-Rapid Amplification of cDNA Ends (RACE) using high-throughput, deep sequencing methods such as Illumina's SBS technology. The degradome encompasses the entire set of proteases that are expressed at a specific time in a given biological material, including tissues, cells, organisms, and biofluids. Thus, sequencing this degradome offers a method for studying and researching the process of RNA degradation. This process is used to identify and quantify RNA degradation products, or fragments, present in any given biological sample. This approach allows for the systematic identification of targets of RNA decay and provides insight into the dynamics of transcriptional and post-transcriptional gene regulation.

miRTarBase is a curated database of MicroRNA-Target Interactions. As a database, miRTarBase has accumulated more than fifty thousand miRNA-target interactions (MTIs), which are collected by manually surveying pertinent literature after data mining of the text systematically to filter research articles related to functional studies of miRNAs. Generally, the collected MTIs are validated experimentally by reporter assay, western blot, microarray and next-generation sequencing experiments. While containing the largest amount of validated MTIs, the miRTarBase provides the most updated collection by comparing with other similar, previously developed databases.
MicroRNA sequencing (miRNA-seq), a type of RNA-Seq, is the use of next-generation sequencing or massively parallel high-throughput DNA sequencing to sequence microRNAs, also called miRNAs. miRNA-seq differs from other forms of RNA-seq in that input material is often enriched for small RNAs. miRNA-seq allows researchers to examine tissue-specific expression patterns, disease associations, and isoforms of miRNAs, and to discover previously uncharacterized miRNAs. Evidence that dysregulated miRNAs play a role in diseases such as cancer has positioned miRNA-seq to potentially become an important tool in the future for diagnostics and prognostics as costs continue to decrease. Like other miRNA profiling technologies, miRNA-Seq has both advantages and disadvantages.
In molecular biology mir-390 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-396 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.











