Related Research Articles

A chromosome is a long DNA molecule with part or all of the genetic material of an organism. In most chromosomes the very long thin DNA fibers are coated with packaging proteins; in eukaryotic cells the most important of these proteins are the histones. These proteins, aided by chaperone proteins, bind to and condense the DNA molecule to maintain its integrity. These chromosomes display a complex three-dimensional structure, which plays a significant role in transcriptional regulation.

The centromere links a pair of sister chromatids together during cell division. This constricted region of chromosome connects the sister chromatids, creating a short arm (p) and a long arm (q) on the chromatids. During mitosis, spindle fibers attach to the centromere via the kinetochore.
Heterochromatin is a tightly packed form of DNA or condensed DNA, which comes in multiple varieties. These varieties lie on a continuum between the two extremes of constitutive heterochromatin and facultative heterochromatin. Both play a role in the expression of genes. Because it is tightly packed, it was thought to be inaccessible to polymerases and therefore not transcribed; however, according to Volpe et al. (2002), and many other papers since, much of this DNA is in fact transcribed, but it is continuously turned over via RNA-induced transcriptional silencing (RITS). Recent studies with electron microscopy and OsO4 staining reveal that the dense packing is not due to the chromatin.
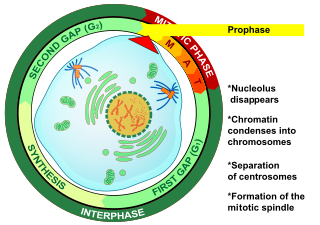
Prophase is the first stage of cell division in both mitosis and meiosis. Beginning after interphase, DNA has already been replicated when the cell enters prophase. The main occurrences in prophase are the condensation of the chromatin reticulum and the disappearance of the nucleolus.

A karyotype is the general appearance of the complete set of chromosomes in the cells of a species or in an individual organism, mainly including their sizes, numbers, and shapes. Karyotyping is the process by which a karyotype is discerned by determining the chromosome complement of an individual, including the number of chromosomes and any abnormalities.

A couple of homologous chromosomes, or homologs, are a set of one maternal and one paternal chromosome that pair up with each other inside a cell during fertilization. Homologs have the same genes in the same loci, where they provide points along each chromosome that enable a pair of chromosomes to align correctly with each other before separating during meiosis. This is the basis for Mendelian inheritance, which characterizes inheritance patterns of genetic material from an organism to its offspring parent developmental cell at the given time and area.

Robertsonian translocation (ROB) is a chromosomal abnormality where the entire long arms of two different chromosomes become fused to each other. It is the most common form of chromosomal translocation in humans, affecting 1 out of every 1,000 babies born. It does not usually cause medical problems, however such individuals are almost always infertile because they are unable to produce gametes with the correct number of chromosomes. In rare cases this translocation results in Down syndrome and Patau syndrome. Robertsonian translocations result in a reduction in the number of chromosomes. A Robertsonian evolutionary fusion, which may have occurred in the common ancestor of humans and other great apes, is the reason humans have 46 chromosomes while all other primates have 48. Detailed DNA studies of chimpanzee, orangutan, gorilla and bonobo apes has determined that where human chromosome 2 is present in our DNA in all four great apes this is split into two separate chromosomes typically numbered 2a and 2b. Similarly, the fact that horses have 64 chromosomes and donkeys 62, and that they can still have common, albeit usually infertile, offspring, may be due to a Robertsonian evolutionary fusion at some point in the descent of today's donkeys from their common ancestor.

Polytene chromosomes are large chromosomes which have thousands of DNA strands. They provide a high level of function in certain tissues such as salivary glands of insects.

An isochromosome is an unbalanced structural abnormality in which the arms of the chromosome are mirror images of each other. The chromosome consists of two copies of either the long (q) arm or the short (p) arm because isochromosome formation is equivalent to a simultaneous duplication and deletion of genetic material. Consequently, there is partial trisomy of the genes present in the isochromosome and partial monosomy of the genes in the lost arm.
Subtelomeres are segments of DNA between telomeric caps and chromatin.

In genetics, a locus is a specific, fixed position on a chromosome where a particular gene or genetic marker is located. Each chromosome carries many genes, with each gene occupying a different position or locus; in humans, the total number of protein-coding genes in a complete haploid set of 23 chromosomes is estimated at 19,000–20,000.
Meiotic drive is a type of intragenomic conflict, whereby one or more loci within a genome will affect a manipulation of the meiotic process in such a way as to favor the transmission of one or more alleles over another, regardless of its phenotypic expression. More simply, meiotic drive is when one copy of a gene is passed on to offspring more than the expected 50% of the time. According to Buckler et al., "Meiotic drive is the subversion of meiosis so that particular genes are preferentially transmitted to the progeny. Meiotic drive generally causes the preferential segregation of small regions of the genome".
An acentric fragment is a segment of a chromosome that lacks a centromere.
A dicentric chromosome is an abnormal chromosome with two centromeres. It is formed through the fusion of two chromosome segments, each with a centromere, resulting in the loss of acentric fragments and the formation of dicentric fragments. The formation of dicentric chromosomes has been attributed to genetic processes, such as Robertsonian translocation and paracentric inversion. Dicentric chromosomes have important roles in the mitotic stability of chromosomes and the formation of pseudodicentric chromosomes. Their existence has been linked to certain natural phenomena such as irradiation and have been documented to underlie certain clinical syndromes, notably Kabuki syndrome. The formation of dicentric chromosomes and their implications on centromere function are studied in certain clinical cytogenetics laboratories.
A chromosomal abnormality, chromosomal anomaly, chromosomal aberration, chromosomal mutation, or chromosomal disorder is a missing, extra, or irregular portion of chromosomal DNA. These can occur in the form of numerical abnormalities, where there is an atypical number of chromosomes, or as structural abnormalities, where one or more individual chromosomes are altered. Chromosome mutation was formerly used in a strict sense to mean a change in a chromosomal segment, involving more than one gene. Chromosome anomalies usually occur when there is an error in cell division following meiosis or mitosis. Chromosome abnormalities may be detected or confirmed by comparing an individual's karyotype, or full set of chromosomes, to a typical karyotype for the species via genetic testing.

Neocentromeres are new centromeres that form at a place on the chromosome that is usually not centromeric. They typically arise due to disruption of the normal centromere. These neocentromeres should not be confused with “knobs”, which were also described as “neocentromeres” in maize in the 1950s. Unlike most normal centromeres, neocentromeres do not contain satellite sequences that are highly repetitive but instead consist of unique sequences. Despite this, most neocentromeres are still able to carry out the functions of normal centromeres in regulating chromosome segregation and inheritance. This raises many questions on what is necessary versus what is sufficient for constituting a centromere.

Robin Campbell Allshire is Professor of Chromosome Biology at University of Edinburgh and a Wellcome Trust Principal Research Fellow. His research group at the Wellcome Trust Centre for Cell Biology focuses on the epigenetic mechanisms governing the assembly of specialised domains of chromatin and their transmission through cell division.
Polycentric is an English adjective, meaning "having more than one center," derived from the Greek words polús ("many") and kentrikós ("center"). Polycentricism is the abstract noun formed from polycentric. They may refer to:
Holocentric chromosomes are chromosomes that possess multiple kinetochores along their length rather than the single centromere typical of other chromosomes. They were first described in cytogenetic experiments in 1935. Since this first observation, the term holocentric chromosome has referred to chromosomes that: i) lack the primary constriction corresponding to the centromere observed in monocentric chromosomes; and ii) possess multiple kinetochores dispersed along the entire chromosomal axis, such that microtubules bind to the chromosome along its entire length and move broadside to the pole from the metaphase plate. Holocentric chromosomes are also termed holokinetic, because, during cell division, the sister chromatids move apart in parallel and do not form the classical V-shaped figures typical of monocentric chromosomes.

The monocentric chromosome is a chromosome that has only one centromere in a chromosome and forms a narrow constriction.
References
- ↑ Neumann P, Navrátilová A, Schroeder-Reiter E, Koblížková A, Steinbauerová V, Chocholová E, et al. (2012-06-21). "Stretching the rules: monocentric chromosomes with multiple centromere domains". PLOS Genetics. 8 (6): e1002777. doi: 10.1371/journal.pgen.1002777 . PMC 3380829 . PMID 22737088.
- ↑ Godward MB (April 1954). "The 'Diffuse' Centromere or Polycentric Chromosomes in Spirogyra". Annals of Botany. 18 (2): 143–144. doi:10.1093/oxfordjournals.aob.a083387.
- ↑ D. Lewis (1986). "Leonard Francis La Cour 28 July 1907–3 November 1984". Biographical Memoirs of Fellows of the Royal Society . 32 (32): 368. doi: 10.1098/rsbm.1986.0011 .