
An oncogene is a gene that has the potential to cause cancer. In tumor cells, these genes are often mutated, or expressed at high levels.

In biology, epigenetics is the study of heritable traits, or a stable change of cell function, that happen without changes to the DNA sequence. The Greek prefix epi- in epigenetics implies features that are "on top of" or "in addition to" the traditional genetic mechanism of inheritance. Epigenetics usually involves a change that is not erased by cell division, and affects the regulation of gene expression. Such effects on cellular and physiological phenotypic traits may result from environmental factors, or be part of normal development. They can lead to cancer.

Metastasis is a pathogenic agent's spread from an initial or primary site to a different or secondary site within the host's body; the term is typically used when referring to metastasis by a cancerous tumor. The newly pathological sites, then, are metastases (mets). It is generally distinguished from cancer invasion, which is the direct extension and penetration by cancer cells into neighboring tissues.

Carcinoma is a malignancy that develops from epithelial cells. Specifically, a carcinoma is a cancer that begins in a tissue that lines the inner or outer surfaces of the body, and that arises from cells originating in the endodermal, mesodermal or ectodermal germ layer during embryogenesis.
Malignant transformation is the process by which cells acquire the properties of cancer. This may occur as a primary process in normal tissue, or secondarily as malignant degeneration of a previously existing benign tumor.
MammaPrint is a prognostic and predictive diagnostic test for early stage breast cancer patients that assess the risk that a tumor will metastasize to other parts of the body. It gives a binary result, high-risk or low-risk classification, and helps physicians determine whether or not a patient will benefit from chemotherapy. Women with a low risk result can safely forego chemotherapy without decreasing likelihood of disease free survival. MammaPrint is part of the personalized medicine portfolio marketed by Agendia.
In medicine, a biomarker is a measurable indicator of the severity or presence of some disease state. It may be defined as a "cellular, biochemical or molecular alteration in cells, tissues or fluids that can be measured and evaluated to indicate normal biological processes, pathogenic processes, or pharmacological responses to a therapeutic intervention." More generally a biomarker is anything that can be used as an indicator of a particular disease state or some other physiological state of an organism. According to the WHO, the indicator may be chemical, physical, or biological in nature - and the measurement may be functional, physiological, biochemical, cellular, or molecular.

Oncogenomics is a sub-field of genomics that characterizes cancer-associated genes. It focuses on genomic, epigenomic and transcript alterations in cancer.

Methylated-DNA--protein-cysteine methyltransferase(MGMT), also known as O6-alkylguanine DNA alkyltransferaseAGT, is a protein that in humans is encoded by the MGMT gene. MGMT is crucial for genome stability. It repairs the naturally occurring mutagenic DNA lesion O6-methylguanine back to guanine and prevents mismatch and errors during DNA replication and transcription. Accordingly, loss of MGMT increases the carcinogenic risk in mice after exposure to alkylating agents. The two bacterial isozymes are Ada and Ogt.
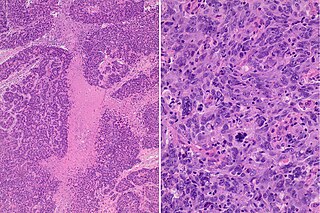
The basal-like carcinoma is a recently proposed subtype of breast cancer defined by its gene expression and protein expression profile.
A metastasis suppressor is a protein that acts to slow or prevent metastases from spreading in the body of an organism with cancer. Metastasis is one of the most lethal cancer processes. This process is responsible for about ninety percent of human cancer deaths. Proteins that act to slow or prevent metastases are different from those that act to suppress tumor growth. Genes for about a dozen such proteins are known in humans and other animals.
Breast cancer classification divides breast cancer into categories according to different schemes criteria and serving a different purpose. The major categories are the histopathological type, the grade of the tumor, the stage of the tumor, and the expression of proteins and genes. As knowledge of cancer cell biology develops these classifications are updated.
A gene signature or gene expression signature is a single or combined group of genes in a cell with a uniquely characteristic pattern of gene expression that occurs as a result of an altered or unaltered biological process or pathogenic medical condition. This is not to be confused with the concept of gene expression profiling. Activating pathways in a regular physiological process or a physiological response to a stimulus results in a cascade of signal transduction and interactions that elicit altered levels of gene expression, which is classified as the gene signature of that physiological process or response. The clinical applications of gene signatures breakdown into prognostic, diagnostic and predictive signatures. The phenotypes that may theoretically be defined by a gene expression signature range from those that predict the survival or prognosis of an individual with a disease, those that are used to differentiate between different subtypes of a disease, to those that predict activation of a particular pathway. Ideally, gene signatures can be used to select a group of patients for whom a particular treatment will be effective.
Cancer is a category of disease characterized by uncontrolled cell growth and proliferation. For cancer to develop, genes regulating cell growth and differentiation must be altered; these mutations are then maintained through subsequent cell divisions and are thus present in all cancerous cells. Gene expression profiling is a technique used in molecular biology to query the expression of thousands of genes simultaneously. In the context of cancer, gene expression profiling has been used to more accurately classify tumors. The information derived from gene expression profiling often helps in predicting the patient's clinical outcome.

Yippee-like 3 (Drosophila) is a protein that in humans is encoded by the YPEL3 gene. YPEL3 has growth inhibitory effects in normal and tumor cell lines. One of five family members (YPEL1-5), YPEL3 was named in reference to its Drosophila melanogaster orthologue. Initially discovered in a gene expression profiling assay of p53 activated MCF7 cells, induction of YPEL3 has been shown to trigger permanent growth arrest or cellular senescence in certain human normal and tumor cell types. DNA methylation of a CpG island near the YPEL3 promoter as well as histone acetylation may represent possible epigenetic mechanisms leading to decreased gene expression in human tumors.

A cancer biomarker refers to a substance or process that is indicative of the presence of cancer in the body. A biomarker may be a molecule secreted by a tumor or a specific response of the body to the presence of cancer. Genetic, epigenetic, proteomic, glycomic, and imaging biomarkers can be used for cancer diagnosis, prognosis, and epidemiology. Ideally, such biomarkers can be assayed in non-invasively collected biofluids like blood or serum.

Cancer epigenetics is the study of epigenetic modifications to the DNA of cancer cells that do not involve a change in the nucleotide sequence, but instead involve a change in the way the genetic code is expressed. Epigenetic mechanisms are necessary to maintain normal sequences of tissue specific gene expression and are crucial for normal development. They may be just as important, if not even more important, than genetic mutations in a cell's transformation to cancer. The disturbance of epigenetic processes in cancers, can lead to a loss of expression of genes that occurs about 10 times more frequently by transcription silencing than by mutations. As Vogelstein et al. points out, in a colorectal cancer there are usually about 3 to 6 driver mutations and 33 to 66 hitchhiker or passenger mutations. However, in colon tumors compared to adjacent normal-appearing colonic mucosa, there are about 600 to 800 heavily methylated CpG islands in the promoters of genes in the tumors while these CpG islands are not methylated in the adjacent mucosa. Manipulation of epigenetic alterations holds great promise for cancer prevention, detection, and therapy. In different types of cancer, a variety of epigenetic mechanisms can be perturbed, such as the silencing of tumor suppressor genes and activation of oncogenes by altered CpG island methylation patterns, histone modifications, and dysregulation of DNA binding proteins. There are several medications which have epigenetic impact, that are now used in a number of these diseases.

J. William Harbour is an American ophthalmologist, ocular oncologist and cancer researcher. He is Chair of the Department of Ophthalmology at the University of Texas Southwestern Medical Center in Dallas. He previously served as the vice chair and director of ocular oncology at the Bascom Palmer Eye Institute and associate director for basic science at the Sylvester Comprehensive Cancer Center of the University of Miami's Miller School of Medicine.
DNA methylation in cancer plays a variety of roles, helping to change the healthy cells by regulation of gene expression to a cancer cells or a diseased cells disease pattern. One of the most widely studied DNA methylation dysregulation is the promoter hypermethylation where the CPGs islands in the promoter regions are methylated contributing or causing genes to be silenced.
CpG island hypermethylation is a phenomenon that is important for the regulation of gene expression in cancer cells, as an epigenetic control aberration responsible for gene inactivation. Hypermethylation of CpG islands has been described in almost every type of tumor.










