
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity.
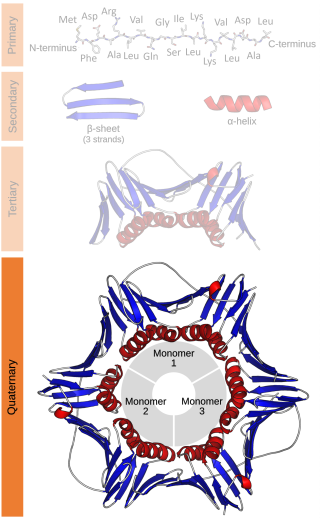
Protein quaternary structure is the fourth classification level of protein structure. Protein quaternary structure refers to the structure of proteins which are themselves composed of two or more smaller protein chains. Protein quaternary structure describes the number and arrangement of multiple folded protein subunits in a multi-subunit complex. It includes organizations from simple dimers to large homooligomers and complexes with defined or variable numbers of subunits. In contrast to the first three levels of protein structure, not all proteins will have a quaternary structure since some proteins function as single units. Protein quaternary structure can also refer to biomolecular complexes of proteins with nucleic acids and other cofactors.

Structural biology, as defined by the Journal of Structural Biology, deals with structural analysis of living material at every level of organization.

Proteomics is the large-scale study of proteins. Proteins are vital macromolecules of all living organisms, with many functions such as the formation of structural fibers of muscle tissue, enzymatic digestion of food, or synthesis and replication of DNA. In addition, other kinds of proteins include antibodies that protect an organism from infection, and hormones that send important signals throughout the body.
The Protein Data Bank (PDB) is a database for the three-dimensional structural data of large biological molecules such as proteins and nucleic acids, which is overseen by the Worldwide Protein Data Bank (wwPDB). These structural data are obtained and deposited by biologists and biochemists worldwide through the use of experimental methodologies such as X-ray crystallography, NMR spectroscopy, and, increasingly, cryo-electron microscopy. All submitted data are reviewed by expert biocurators and, once approved, are made freely available on the Internet under the CC0 Public Domain Dedication. Global access to the data is provided by the websites of the wwPDB member organisations.

A biomolecule or biological molecule is loosely defined as a molecule produced by a living organism and essential to one or more typically biological processes. Biomolecules include large macromolecules such as proteins, carbohydrates, lipids, and nucleic acids, as well as small molecules such as vitamins and hormones. A general name for this class of material is biological materials. Biomolecules are an important element of living organisms. They are often endogenous, i.e. produced within the organism, but organisms usually also need exogenous biomolecules, for example certain nutrients, to survive.

In biochemistry and molecular biology, a binding site is a region on a macromolecule such as a protein that binds to another molecule with specificity. The binding partner of the macromolecule is often referred to as a ligand. Ligands may include other proteins, enzyme substrates, second messengers, hormones, or allosteric modulators. The binding event is often, but not always, accompanied by a conformational change that alters the protein's function. Binding to protein binding sites is most often reversible, but can also be covalent reversible or irreversible.

The enzyme cholinesterase (EC 3.1.1.8, choline esterase; systematic name acylcholine acylhydrolase) catalyses the hydrolysis of choline-based esters:

In biochemistry, a Ramachandran plot, originally developed in 1963 by G. N. Ramachandran, C. Ramakrishnan, and V. Sasisekharan, is a way to visualize energetically allowed regions for backbone dihedral angles ψ against φ of amino acid residues in protein structure. The figure on the left illustrates the definition of the φ and ψ backbone dihedral angles. The ω angle at the peptide bond is normally 180°, since the partial-double-bond character keeps the peptide bond planar. The figure in the top right shows the allowed φ,ψ backbone conformational regions from the Ramachandran et al. 1963 and 1968 hard-sphere calculations: full radius in solid outline, reduced radius in dashed, and relaxed tau (N-Cα-C) angle in dotted lines. Because dihedral angle values are circular and 0° is the same as 360°, the edges of the Ramachandran plot "wrap" right-to-left and bottom-to-top. For instance, the small strip of allowed values along the lower-left edge of the plot are a continuation of the large, extended-chain region at upper left.

The cytochrome b6f complex (plastoquinol/plastocyanin reductase or plastoquinol/plastocyanin oxidoreductase; EC 7.1.1.6) is an enzyme found in the thylakoid membrane in chloroplasts of plants, cyanobacteria, and green algae, that catalyzes the transfer of electrons from plastoquinol to plastocyanin:

Huperzine A is a naturally-occurring sesquiterpene alkaloid compound found in the firmoss Huperzia serrata and in varying quantities in other food Huperzia species, including H. elmeri, H. carinat, and H. aqualupian. Huperzine A has been investigated as a treatment for neurological conditions such as Alzheimer's disease, but a 2013 meta-analysis of those studies concluded that they were of poor methodological quality and the findings should be interpreted with caution. Huperzine A inhibits the breakdown of the neurotransmitter acetylcholine (ACh) by the enzyme acetylcholinesterase. It is also an antagonist of the NMDA-receptor. It is commonly available over the counter as a nutritional supplement and marketed as a memory and concentration enhancer.

Joel L. Sussman is an Israeli crystallographer best known for his studies on acetylcholinesterase, a key protein involved in transmission of nerve signals. He is Professor Emeritus of Structural Biology at the Weizmann Institute of Science in Rehovot and is Co-Director of the Israel Structural Proteomics Center.

Jmol is computer software for molecular modelling of chemical structures in 3 dimensions. It is an open-source Java viewer for chemical structures in 3D. The name originated from [J]ava + [mol]ecules, and also the mol file format.

Acetylcholinesterase (HGNC symbol ACHE; EC 3.1.1.7; systematic name acetylcholine acetylhydrolase), also known as AChE, AChase or acetylhydrolase, is the primary cholinesterase in the body. It is an enzyme that catalyzes the breakdown of acetylcholine and some other choline esters that function as neurotransmitters:

The Protein Structure Initiative (PSI) was a USA based project that aimed at accelerating discovery in structural genomics and contribute to understanding biological function. Funded by the U.S. National Institute of General Medical Sciences (NIGMS) between 2000 and 2015, its aim was to reduce the cost and time required to determine three-dimensional protein structures and to develop techniques for solving challenging problems in structural biology, including membrane proteins. Over a dozen research centers have been supported by the PSI for work in building and maintaining high-throughput structural genomics pipelines, developing computational protein structure prediction methods, organizing and disseminating information generated by the PSI, and applying high-throughput structure determination to study a broad range of important biological and biomedical problems.
PDBWiki was a wiki that functioned as a user-contributed database of protein structure annotations, listing all the protein structures available in the Protein Data Bank (PDB). It ran on the MediaWiki wiki application from 2007 to 2013. The website went offline in 2014 and there has not been any way to subsequently access the information that was contributed. PDBWiki contained details of more than 50,000 protein structures and over 50 'user-contributed' annotations, making it a significant resource for the structural biology community.
In biology, a protein structure database is a database that is modeled around the various experimentally determined protein structures. The aim of most protein structure databases is to organize and annotate the protein structures, providing the biological community access to the experimental data in a useful way. Data included in protein structure databases often includes three-dimensional coordinates as well as experimental information, such as unit cell dimensions and angles for x-ray crystallography determined structures. Though most instances, in this case either proteins or a specific structure determinations of a protein, also contain sequence information and some databases even provide means for performing sequence based queries, the primary attribute of a structure database is structural information, whereas sequence databases focus on sequence information, and contain no structural information for the majority of entries. Protein structure databases are critical for many efforts in computational biology such as structure based drug design, both in developing the computational methods used and in providing a large experimental dataset used by some methods to provide insights about the function of a protein.

In molecular biology, the term macromolecular assembly (MA) refers to massive chemical structures such as viruses and non-biologic nanoparticles, cellular organelles and membranes and ribosomes, etc. that are complex mixtures of polypeptide, polynucleotide, polysaccharide or other polymeric macromolecules. They are generally of more than one of these types, and the mixtures are defined spatially, and with regard to their underlying chemical composition and structure. Macromolecules are found in living and nonliving things, and are composed of many hundreds or thousands of atoms held together by covalent bonds; they are often characterized by repeating units. Assemblies of these can likewise be biologic or non-biologic, though the MA term is more commonly applied in biology, and the term supramolecular assembly is more often applied in non-biologic contexts. MAs of macromolecules are held in their defined forms by non-covalent intermolecular interactions, and can be in either non-repeating structures, or in repeating linear, circular, spiral, or other patterns. The process by which MAs are formed has been termed molecular self-assembly, a term especially applied in non-biologic contexts. A wide variety of physical/biophysical, chemical/biochemical, and computational methods exist for the study of MA; given the scale of MAs, efforts to elaborate their composition and structure and discern mechanisms underlying their functions are at the forefront of modern structure science.

Fasciculins are a class of toxic proteins found in certain snake venoms, notably some species of mamba. Investigations have revealed distinct forms in some green mamba venoms, in particular FAS1 and FAS2 Fasciculins are so called because they cause intense fasciculation in muscle fascicles of susceptible organisms, such as the preferred prey of the snakes. This effect helps to incapacitate the muscles, either killing the prey, or paralysing it so that the snake can swallow it.

Cryogenic electron microscopy (cryo-EM) is a cryomicroscopy technique applied on samples cooled to cryogenic temperatures. For biological specimens, the structure is preserved by embedding in an environment of vitreous ice. An aqueous sample solution is applied to a grid-mesh and plunge-frozen in liquid ethane or a mixture of liquid ethane and propane. While development of the technique began in the 1970s, recent advances in detector technology and software algorithms have allowed for the determination of biomolecular structures at near-atomic resolution. This has attracted wide attention to the approach as an alternative to X-ray crystallography or NMR spectroscopy for macromolecular structure determination without the need for crystallization.















