In biology, quorum sensing is the ability to detect and to respond to cell population density by gene regulation. As one example, quorum sensing (QS) enables bacteria to restrict the expression of specific genes to the high cell densities at which the resulting phenotypes will be most beneficial. Many species of bacteria use quorum sensing to coordinate gene expression according to the density of their local population. In similar fashion, some social insects use quorum sensing to determine where to nest.
The gene rpoS encodes the sigma factor sigma-38, a 37.8 kD protein in Escherichia coli. Sigma factors are proteins that regulate transcription in bacteria. Sigma factors can be activated in response to different environmental conditions. rpoS is transcribed in late exponential phase, and RpoS is the primary regulator of stationary phase genes. RpoS is a central regulator of the general stress response and operates in both a retroactive and a proactive manner: it not only allows the cell to survive environmental challenges, but it also prepares the cell for subsequent stresses (cross-protection). The transcriptional regulator CsgD is central to biofilm formation, controlling the expression of the curli structural and export proteins, and the diguanylate cyclase, adrA, which indirectly activates cellulose production. The rpoS gene most likely originated in the gammaproteobacteria.
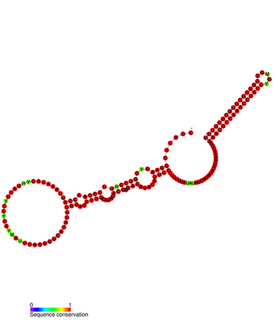
The PrrB/RsmZ RNA family are a group of related non-coding RNA molecules found in bacteria. PrrB RNA is able to phenotypically complement gacS and gacA mutants and is itself regulated by the GacS-GacA two-component signal transduction system. Inactivation of the prrB gene in Pseudomonas fluorescens F113 resulted in a significant reduction of 2, 4-diacetylphloroglucinol (Phl) and hydrogen cyanide (HCN) production, while increased metabolite production was observed when prrB was overexpressed. The prrB gene sequence contains a number of imperfect repeats of the consensus sequence 5'-AGGA-3', and sequence analysis predicted a complex secondary structure featuring multiple putative stem-loops with the consensus sequences predominantly positioned at the single-stranded regions at the ends of the stem-loops. This structure is similar to the CsrB and RsmB regulatory RNAs, suggesting this RNA also interacts with a CsrA-like protein.

The micF RNA is a non-coding RNA stress response gene found in Escherichia coli and related bacteria that post-transcriptionally controls expression of the outer membrane porin gene ompF. The micF gene encodes a non-translated 93 nucleotide antisense RNA that binds its target ompF mRNA and regulates ompF expression by inhibiting translation and inducing degradation of the message. In addition, other factors, such as the RNA chaperone protein StpA also play a role in this regulatory system. Expression of micF is controlled by both environmental and internal stress factors. Four transcriptional regulators are known to bind the micF promoter region and activate micF expression.

The repression of heat shock gene expression (ROSE) element is an RNA element found in the 5' UTR of some heat shock protein's mRNAs. The ROSE element is an RNA thermometer that negatively regulates heat shock gene expression. The secondary structure is thought to be altered by temperature, thus it is an RNA thermometer. This structure blocks access to the ribosome binding site at normal temperatures. During heat shock however, the structure changes freeing the ribosome binding site and allowing expression to occur.
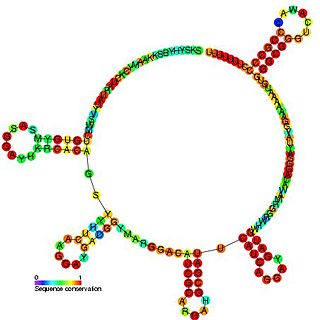
RNAIII is a small RNA which is known to regulate the expression of many Staphylococcus aureus genes encoding exoproteins and cell wall associated proteins. In S. aureus, RNAIII acts as the effector of the agr quorum sensing system and is transcribed from the P3 operon. The RNAIII transcript also contains the 26 amino acid delta-haemolysin gene (hld). RNAIII regulates the expression of the transcription factor rot by blocking its translation. It has been suggested that RNAIII binds to the rot mRNA in an antisense fashion occluding the Shine-Dalgarno sequence.

The rsmY RNA family is a set of related non-coding RNA genes, that like RsmZ, is regulated by the GacS/GacA signal transduction system in the plant-beneficial soil bacterium and biocontrol model organism Pseudomonas fluorescens CHA0. GacA/GacS target genes are translationally repressed by the small RNA binding protein RsmA. RsmY and RsmZ RNAs bind RsmA to relieve this repression and so enhance secondary metabolism and biocontrol traits.

RyhB RNA is a 90 nucleotide RNA that down-regulates a set of iron-storage and iron-using proteins when iron is limiting; it is itself negatively regulated by the ferric uptake repressor protein, Fur.
Autoinducers are signaling molecules that are produced in response to changes in cell-population density. As the density of quorum sensing bacterial cells increases so does the concentration of the autoinducer. Detection of signal molecules by bacteria acts as stimulation which leads to altered gene expression once the minimal threshold is reached. Quorum sensing is a phenomenon that allows both Gram-negative and Gram-positive bacteria to sense one another and to regulate a wide variety of physiological activities. Such activities include symbiosis, virulence, motility, antibiotic production, and biofilm formation. Autoinducers come in a number of different forms depending on the species, but the effect that they have is similar in many cases. Autoinducers allow bacteria to communicate both within and between different species. This communication alters gene expression and allows bacteria to mount coordinated responses to their environments, in a manner that is comparable to behavior and signaling in higher organisms. Not surprisingly, it has been suggested that quorum sensing may have been an important evolutionary milestone that ultimately gave rise to multicellular life forms.

Lactonase is a metalloenzyme, produced by certain species of bacteria, which targets and inactivates acylated homoserine lactones (AHLs).
Pseudomonas sRNA are non-coding RNAs (ncRNA) that were predicted by the bioinformatic program SRNApredict2. This program identifies putative sRNAs by searching for co-localization of genetic features commonly associated with sRNA-encoding genes and the expression of the predicted sRNAs was subsequently confirmed by Northern blot analysis. These sRNAs have been shown to be conserved across several pseudomonas species but their function is yet to be determined. Using Tet-Trap genetic approach RNAT genes post-transcriptionally regulated by temperature upshift were identified: ptxS and PA5194.

The gabT RNA motif is the name of a conserved RNA structure identified by bioinformatics whose function is unknown. The gabT motif has been detected exclusively in bacteria within the genus Pseudomonas, and is found only upstream of gabT genes, and downstream to gabD genes.

The Pseudomon-1 RNA motif is a conserved RNA identified by bioinformatics. It is used by most species whose genomes have been sequenced and that are classified within the genus Pseudomonas, and is also present in Azotobacter vinelandii, a closely related species. It is presumed to function as a non-coding RNA. Pseudomon-1 RNAs consistently have a downstream rho-independent transcription terminator.
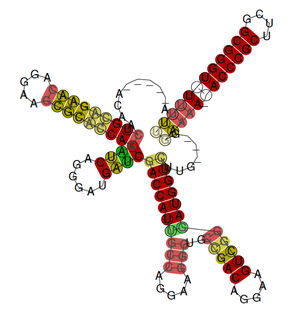
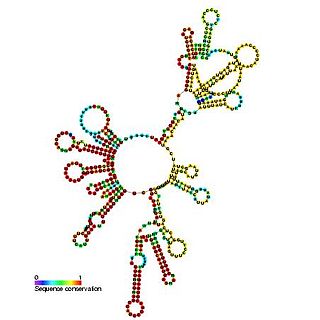
The rsmX gene is part of the Rsm/Csr family of non-coding RNAs (ncRNAs). Members of the Rsm/Csr family are present in a diverse range of bacteria, including Escherichia coli, Erwinia, Salmonella, Vibrio and Pseudomonas. These ncRNAs act by sequestering translational repressor proteins, called RsmA, activating expression of downstream genes that would normally be blocked by the repressors. Sequestering of target proteins is dependent upon exposed GGA motifs in the stem loops of the ncRNAs. Typically, the activated genes are involved in secondary metabolism, biofilm formation and motility.
Bacterial small RNAs (sRNA) are small RNAs produced by bacteria; they are 50- to 500-nucleotide non-coding RNA molecules, highly structured and containing several stem-loops. Numerous sRNAs have been identified using both computational analysis and laboratory-based techniques such as Northern blotting, microarrays and RNA-Seq in a number of bacterial species including Escherichia coli, the model pathogen Salmonella, the nitrogen-fixing alphaproteobacterium Sinorhizobium meliloti, marine cyanobacteria, Francisella tularensis, Streptococcus pyogenes, the pathogen Staphylococcus aureus, and the plant pathogen Xanthomonas oryzae pathovar oryzae. Bacterial sRNAs affect how genes are expressed within bacterial cells via interaction with mRNA or protein, and thus can affect a variety of bacterial functions like metabolism, virulence, environmental stress response, and structure.

In molecular biology, the ferric uptake regulator family is a family of bacterial proteins involved in regulating metal ion uptake and in metal homeostasis. The family is named for its founding member, known as the ferric uptake regulator or ferric uptake regulatory protein (Fur). Fur proteins are responsible for controlling the intracellular concentration of iron in many bacteria. Iron is essential for most organisms, but its concentration must be carefully managed over a wide range of environmental conditions; high concentrations can be toxic due to the formation of reactive oxygen species.
PhrS is a bacterial small RNA found in Pseudomonas aeruginosa. It was first identified in a RNAomics screen and has since been found to act as a link between oxygen availability and quorum sensing.
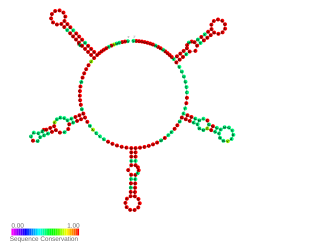
NrsZ is a bacterial small RNA found in the opportunistic pathogen Pseudomonas aeruginosa PAO1. Its transcription is induced during nitrogen limitation by the NtrB/C two-component system together with the alternative sigma factor RpoN. NrsZ by activating rhlA positively regulates the production of rhamnolipid surfactants needed for swarming motility.
Laurence G. Rahmeis Professor of Surgery and Microbiology at Harvard Medical School (HMS). At Massachusetts General Hospital (MGH) she also holds the title of Director of the Molecular Surgical Laboratory as a Microbiologists in the Department of Surgery and Molecular Biology at. Additionally, she hols a Senior Scientific Staff position at Shriners Hospitals for Children-Boston. She received her B.Sc. from the University of Naples, Italy, her M.S. from the Institute of Genetics and Biophysics C.N.R., University of Naples, Italy, and her Ph.D. from the University of California at Berkeley. She completed her postdoctoral training at the Department of Molecular Biology Massachusetts General Hospital/ and Department of Genetics Harvard Medical School. She was also awarded an honorary M.S. degree from Harvard Medical School. Laurence Rahme has published an extensive number (100+) of scientific articles and holds more than 15 patents with applications to combat bacterial infections and strategies to limit the emergence of antibiotic resistant strains. She has also been on, or currently serves on Advisory and Editorial Boards of numerous scientific journals and served as an ad-hoc member on review panels at the National Institutes of Health (NIH), National Science Foundation (NSF), Department of Defense (DoD) and several national and international research foundations. Her numerous prizes and honors include being elected as an American Academy of Microbiology Fellow in 2017.