
Cyanobacteria, also known as Cyanophyta, are a phylum of Gram-negative bacteria that obtain energy via photosynthesis. The name cyanobacteria comes from their color, giving them their other name, "blue-green algae", though modern botanists restrict the term algae to eukaryotes and do not apply it to cyanobacteria, which are prokaryotes. They appear to have originated in freshwater or a terrestrial environment. Sericytochromatia, the proposed name of the paraphyletic and most basal group, is the ancestor of both the non-photosynthetic group Melainabacteria and the photosynthetic cyanobacteria, also called Oxyphotobacteria.
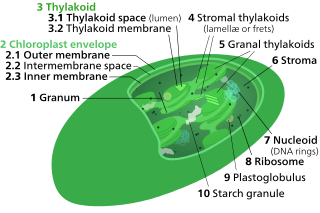
Thylakoids are membrane-bound compartments inside chloroplasts and cyanobacteria. They are the site of the light-dependent reactions of photosynthesis. Thylakoids consist of a thylakoid membrane surrounding a thylakoid lumen. Chloroplast thylakoids frequently form stacks of disks referred to as grana. Grana are connected by intergranal/stromal thylakoids, which join granum stacks together as a single functional compartment.

Photosystem I is one of two photosystems in the photosynthetic light reactions of algae, plants, and cyanobacteria. Photosystem I is an integral membrane protein complex that uses light energy to catalyze the transfer of electrons across the thylakoid membrane from plastocyanin to ferredoxin. Ultimately, the electrons that are transferred by Photosystem I are used to produce the high energy carrier NADPH. The combined action of the entire photosynthetic electron transport chain also produces a proton-motive force that is used to generate ATP. PSI is composed of more than 110 cofactors, significantly more than Photosystem II.

Phycobilisomes are light harvesting antennae of photosystem II in cyanobacteria, red algae and glaucophytes. It was lost in the plastids of green algae/ plants (chloroplasts).
Glutamine synthetase (GS) is an enzyme that plays an essential role in the metabolism of nitrogen by catalyzing the condensation of glutamate and ammonia to form glutamine:
In enzymology, a glucosylglycerol 3-phosphatase (EC 3.1.3.69) is an enzyme that catalyzes the chemical reaction

Photosynthetic reaction centre proteins are main protein components of photosynthetic reaction centres (RCs) of bacteria and plants. They are transmembrane proteins embedded in the chloroplast thylakoid or bacterial cell membrane.
Synechocystis sp. PCC6803 is a strain of unicellular, freshwater cyanobacteria. Synechocystis sp. PCC6803 is capable of both phototrophic growth by oxygenic photosynthesis during light periods and heterotrophic growth by glycolysis and oxidative phosphorylation during dark periods. Gene expression is regulated by a circadian clock and the organism can effectively anticipate transitions between the light and dark phases.

The glutamine riboswitch is a conserved RNA structure that was predicted by bioinformatics. It is present in a variety of lineages of cyanobacteria, as well as some phages that infect cyanobacteria. It is also found in DNA extracted from uncultivated bacteria living in the ocean that are presumably species of cyanobacteria.

PhotoRC RNA motifs refer to conserved RNA structures that are associated with genes acting in the photosynthetic reaction centre of photosynthetic bacteria. Two such RNA classes were identified and called the PhotoRC-I and PhotoRC-II motifs. PhotoRC-I RNAs were detected in the genomes of some cyanobacteria. Although no PhotoRC-II RNA has been detected in cyanobacteria, one is found in the genome of a purified phage that infects cyanobacteria. Both PhotoRC-I and PhotoRC-II RNAs are present in sequences derived from DNA that was extracted from uncultivated marine bacteria.
In molecular biology, the BtpA protein family is a family of proteins which includes BtpA. BtpA appears to play a role in the stabilisation of photosystem I. It is an extrinsic membrane protein located on the cytoplasmic side of the thylakoid membrane. Homologs of BtpA are found in the crenarchaeota and euryarchaeota, where their function remains unknown. The Ycf4 protein is firmly associated with the thylakoid membrane, presumably through a transmembrane domain. Ycf4 co-fractionates with a protein complex larger than PSI upon sucrose density gradient centrifugation of solubilised thylakoids. The Ycf3 protein is loosely associated with the thylakoid membrane and can be released from the membrane with sodium carbonate. This suggests that Ycf3 is not part of a stable complex and that it probably interacts transiently with its partners. Ycf3 contains a number of tetratricopeptide repeats (TPR); TPR is a structural motif present in a wide range of proteins, which mediates protein-protein interactions.
Plastoglobulins is a family of proteins prominent found in lipid globules in plastids of flowering plants. It shows sequence similarities to the PAP/fibrillin family. PGL and similar proteins can be found in most algae, cyanobacteria and plants, but no other life forms; it suggests a role for PGL in oxygenic photosynthesis.
In molecular biology, Cyanobacterial non-coding RNAs are non-coding RNAs which have been identified in species of cyanobacteria. Large scale screens have identified 21 Yfr in the marine cyanobacterium Prochlorococcus and related species such as Synechococcus. These include the Yfr1 and Yfr2 RNAs. In Prochlorococcus and Synechocystis, non-coding RNAs have been shown to regulate gene expression. NsiR4, widely conserved throughout the cyanobacterial phylum, has been shown to be involved in nitrogen assimilation control in Synechocystis sp. PCC 6803 and in the filamentous, nitrogen-fixing Anabaena sp. PCC 7120.
Biological photovoltaics (BPV) is an energy-generating technology which uses oxygenic photoautotrophic organisms, or fractions thereof, to harvest light energy and produce electrical power. Biological photovoltaic devices are a type of biological electrochemical system, or microbial fuel cell, and are sometimes also called photo-microbial fuel cells or “living solar cells”. In a biological photovoltaic system, electrons generated by photolysis of water are transferred to an anode. A relatively high-potential reaction takes place at the cathode, and the resulting potential difference drives current through an external circuit to do useful work. It is hoped that using a living organism as the light harvesting material, will make biological photovoltaics a cost-effective alternative to synthetic light-energy-transduction technologies such as silicon-based photovoltaics.
Ruberedoxin A (RubA) is a protein conserved across all studied oxygenic photoautotrophs.

Cyanothece is a genus of unicellular, diazotrophic, oxygenic photosynthesizing cyanobacteria.
Iron-starvation-induced protein A, also known as isiA, is a photosynthesis-related chlorophyll-containing protein found in cyanobacteria. It belongs to the chlorophyll-a/b-binding family of proteins, and has been shown to have a photoprotection role in preventing oxidative damage via energy dissipation. It was originally identified under Fe starvation, and thus received the name iron-starvation-induced protein A. However, the protein has more recently been found to respond to a variety of stress conditions such as high irradiance. It can aggregate with carotenoids and form rings around the PSI reaction center complexes to aid in photoprotective energy dissipation.
Orange carotenoid protein (OCP) is a water-soluble protein which plays a role in photoprotection in diverse cyanobacteria. It is the only photoactive protein known to use a carotenoid as the photoresponsive chromophore. The protein consists of two domains, with a single keto-carotenoid molecule non-covalently bound between the two domains. It is a very efficient quencher of excitation energy absorbed by the primary light-harvesting antenna complexes of cyanobacteria, the phycobilisomes. The quenching is induced by blue-green light. It is also capable of preventing oxidative damage by directly scavenging singlet oxygen (1O2).

Crocosphaera watsonii is an isolate of a species of unicellular, diazotrophic marine cyanobacteria which represent less than 0.1% of the marine microbial population. They thrive in offshore, open-ocean oligotrophic regions where the waters are warmer than 24 degrees Celsius. Crocosphaera watsonii cell density can exceed 1,000 cells per milliliter within the euphotic zone; however, their growth may be limited by the concentration of phosphorus. Crocosphaera watsonii are able to contribute to the oceanic carbon and nitrogen budgets in tropical oceans due to their size, abundance, and rapid growth rate. Crocosphaera watsonii are unicellular nitrogen fixers that fix atmospheric nitrogen to ammonia during the night and contribute to new nitrogen in the oceans. They are a major source of nitrogen to open-ocean systems. Nitrogen fixation is important in the oceans as it not only allows phytoplankton to continue growing when nitrogen and ammonium are in very low supply but it also replenishes other forms of nitrogen, thus fertilizing the ocean and allowing more phytoplankton growth.
A plastid is a membrane-bound organelle found in plants, algae and other eukaryotic organisms that contribute to the production of pigment molecules. Most plastids are photosynthetic, thus leading to color production and energy storage or production. There are many types of plastids in plants alone, but all plastids can be separated based on the number of times they have undergone endosymbiotic events. Currently there are three types of plastids; primary, secondary and tertiary. Endosymbiosis is reputed to have led to the evolution of eukaryotic organisms today, although the timeline is highly debated.











