
Pseudomonas aeruginosa is a common encapsulated, Gram-negative, aerobic–facultatively anaerobic, rod-shaped bacterium that can cause disease in plants and animals, including humans. A species of considerable medical importance, P. aeruginosa is a multidrug resistant pathogen recognized for its ubiquity, its intrinsically advanced antibiotic resistance mechanisms, and its association with serious illnesses – hospital-acquired infections such as ventilator-associated pneumonia and various sepsis syndromes.

The repression of heat shock gene expression (ROSE) element is an RNA element found in the 5' UTR of some heat shock protein's mRNAs. The ROSE element is an RNA thermometer that negatively regulates heat shock gene expression. The secondary structure is thought to be altered by temperature, thus it is an RNA thermometer. This structure blocks access to the ribosome binding site at normal temperatures. During heat shock however, the structure changes freeing the ribosome binding site and allowing expression to occur.

The SAM riboswitch is found upstream of a number of genes which code for proteins involved in methionine or cysteine biosynthesis in Gram-positive bacteria. Two SAM riboswitches in Bacillus subtilis that were experimentally studied act at the level of transcription termination control. The predicted secondary structure consists of a complex stem-loop region followed by a single stem-loop terminator region. An alternative and mutually exclusive form involves bases in the 3' segment of helix 1 with those in the 5' region of helix 5 to form a structure termed the anti-terminator form. When SAM is unbound, the anti-terminator sequence sequesters the terminator sequence so the terminator is unable to form, allowing the polymerase to read-through the downstream gene. When S-Adenosyl methionine (SAM) is bound to the aptamer, the anti-terminator is sequestered by an anti-anti-terminator; the terminator forms and terminates the transcription. However, many SAM riboswitches are likely to regulate gene expression at the level of translation.

ATP-dependent Clp protease proteolytic subunit (ClpP) is an enzyme that in humans is encoded by the CLPP gene. This protein is an essential component to form the protein complex of Clp protease.

The c4 antisense RNA is a non-coding RNA used by certain phages that infect bacteria. It was initially identified in the P1 and P7 phages of E. coli. The identification of c4 antisense RNAs solved the mystery of the mechanism for regulation of the ant gene, which is an anti-repressor.
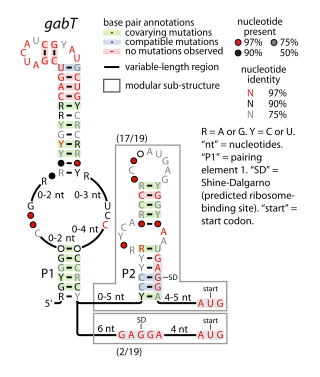
The gabT RNA motif is the name of a conserved RNA structure identified by bioinformatics whose function is unknown. The gabT motif has been detected exclusively in bacteria within the genus Pseudomonas, and is found only upstream of gabT genes, and downstream to gabD genes.

The gyrA RNA motif is a conserved RNA structure identified by bioinformatics. The RNAs are present in multiple species of bacteria within the order Pseudomonadales. This order contains the genus Pseudomonas, which includes the opportunistic human pathogen Pseudomonas aeruginosa and Pseudomonas syringae, a plant pathogen.
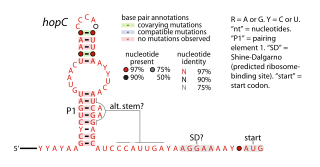
The hopC RNA motif is a predicted cis-regulatory element identified by a bioinformatic screen for conserved RNA secondary structures. hopC RNAs are exclusively found within bacteria classified within the genus Helicobacter, some of which are human pathogens that infect the stomach and can cause ulcers.

The L17 downstream element RNA motif is a conserved RNA structure identified in bacteria by bioinformatics. All known L17 downstream elements were detected immediately downstream of genes encoding the L17 subunit of the ribosome, and therefore might be in the 3' untranslated regions of these genes. The element is found in a variety of lactic acid bacteria and in the genus Listeria.
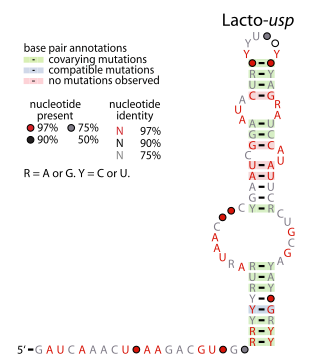
The Lacto-usp RNA motif is a conserved RNA structure identified in bacteria by bioinformatics. Lacto-usp RNAs are found exclusively in lactic acid bacteria, and exclusively in the possible 5′ untranslated regions of operons that contain a hypothetical gene and a usp gene. The usp gene encodes the universal stress protein. It was proposed that the Lacto-usp might correspond to the 6S RNA of the relevant species, because four of five of these species lack a predicted 6S RNA, and 6S RNAs commonly occur in 5′ UTRs of usp genes. However, given that the Lacto-usp RNA motif is much shorter than the standard 6S RNA structure, the function of Lacto-usp RNAs remains unclear.

The livK RNA motif describes a conserved RNA structure that was discovered using bioinformatics. The livK motif is detected only in the species Pseudomonas syringae. It is found in the potential 5' untranslated regions of livK genes and downstream livM and livH genes, as well as the 5' UTRs of amidase genes. The liv genes are predicted to be transporters of branched-chain amino acids, i.e., leucine, isoleucine or valine. The specific reaction catalyzed the amidase genes is not predicted.

The psaA RNA motif describes a class of RNAs with a common secondary structure. psaA RNAs are exclusively found in locations that presumably correspond to the 5' untranslated regions of operons formed of psaA and psaB genes. For this reason, it was hypothesized that psaA RNAs function as cis-regulatory elements of these genes. The psaAB genes encode proteins that form subunits in the photosystem I structure used for photosynthesis. psaA RNAs have been detected only in cyanobacteria, which is consistent with their association with photosynthesis.

The Pseudomon-1 RNA motif is a conserved RNA identified by bioinformatics. It is used by most species whose genomes have been sequenced and that are classified within the genus Pseudomonas, and is also present in Azotobacter vinelandii, a closely related species. It is presumed to function as a non-coding RNA. Pseudomon-1 RNAs consistently have a downstream rho-independent transcription terminator.

The Pseudomon-Rho RNA motif refers to a conserved RNA structure that was discovered using bioinformatics. The RNAs that conform to this motif are found in species within the genus Pseudomonas, as well as the related Azotobacter vinelandii. They are consistently located in what could be the 5' untranslated regions of genes that encode the Rho factor protein, and this arrangement in bacteria suggested that Pseudomon-Rho RNAs might be cis-regulatory elements that regulate concentrations of the Rho protein.

The rmf RNA motif is a conserved RNA structure that was originally detected using bioinformatics. rmf RNAs are consistently foundwithin species classified into the genus Pseudomonas, and is located potentially in the 5′ untranslated regions of rmf genes. These genes encodes the ribosome modulation factor protein, which affects the translation of genes by modifying ribosome structure in response to stress such as starvation. This ribosome modulation is a part of the stringent response in bacteria. The likely biological role of rmf RNAs is ambiguous. Since the RNA could be in the 5′ UTRs of protein-coding genes, it was hypothesized that it functions as a cis-regulatory element. This hypothesis is bolstered by the observation that ribosome modulation factor binds ribosomal RNA, and many cis-regulatory RNAs called ribosomal protein leaders participate in a feedback regulation mechanism by binding to proteins that normally bind to ribosomal RNA. However, since rmf RNAs are not very close to the rmf genes, they might function as non-coding RNAs.

The SAM-Chlorobi RNA motif is a conserved RNA structure that was identified by bioinformatics. The RNAs are found only in bacteria classified as within the phylum Chlorobiota. These RNAs are always in the 5' untranslated regions of operons that contain metK and ahcY genes. metK genes encode methionine adenosyltransferase, which synthesizes S-adenosyl methionine (SAM), and ahcY genes encode S-adenosylhomocysteine hydrolase, which degrade the related metabolite S-Adenosyl-L-homocysteine (SAH). In fact all predicted metK and ahcY genes within Chlorobiota bacteria as of 2010 are preceded by predicted SAM-Chlorobi RNAs. Predicted promoter sequences are consistently found upstream of SAM-Chlorobi RNAs, and these promoter sequences imply that SAM-Chlorobi RNAs are indeed transcribed as RNAs. The promoter sequences are commonly associated with strong transcription in the phyla Chlorobiota and Bacteroidota, but are not used by most lineages of bacteria. The placement of SAM-Chlorobi RNAs suggests that they are involved in the regulation of the metK/ahcY operon through an unknown mechanism.

The sucC RNA motif is a conserved RNA structure discovered using bioinformatics. sucC RNAs are found in the genus Pseudomonas, and are consistently found in possible 5' untranslated regions of sucC genes. These genes encode Succinyl coenzyme A synthetase, and are hypothesised to be regulated by the sucC RNAs. sucC genes participate in the citric acid cycle, and another gene involved in the citric acid cycle, sucA, is also predicted to be regulated by a conserved RNA structure.

The yjdF RNA motif is a conserved RNA structure identified using bioinformatics. Most yjdF RNAs are located in bacteria classified within the phylum Bacillota. A yjdF RNA is found in the presumed 5' untranslated region of the yjdF gene in Bacillus subtilis, and almost all yjdF RNAs are found in the 5' UTRs of homologs of this gene. The function of the yjdF gene is unknown, but the protein that it is predicted to encode is classified by the Pfam Database as DUF2992.

The eps-Associated RNA element is a conserved RNA motif associated with exopolysaccharide (eps) or capsule biosynthesis genes in a subset of bacteria classified within the order Bacillales. It was initially discovered in Bacillus subtilis, located between the second and third gene in the eps operon. Deletion of the EAR element impairs biofilm formation.

The ivy-DE RNA motif is a conserved RNA structure that was discovered by bioinformatics. ivy-DE motifs are found in the genus Pseudomonas.




















