Related Research Articles
Molecular biology is a branch of biology that seeks to understand the molecular basis of biological activity in and between cells, including biomolecular synthesis, modification, mechanisms, and interactions.
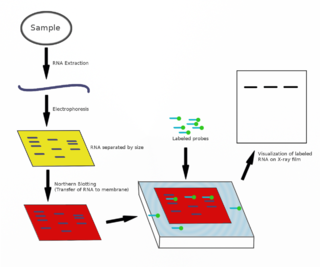
The northern blot, or RNA blot, is a technique used in molecular biology research to study gene expression by detection of RNA in a sample.

The polymerase chain reaction (PCR) is a method widely used to make millions to billions of copies of a specific DNA sample rapidly, allowing scientists to amplify a very small sample of DNA sufficiently to enable detailed study. PCR was invented in 1983 by American biochemist Kary Mullis at Cetus Corporation. Mullis and biochemist Michael Smith, who had developed other essential ways of manipulating DNA, were jointly awarded the Nobel Prize in Chemistry in 1993.

Reverse transcription polymerase chain reaction (RT-PCR) is a laboratory technique combining reverse transcription of RNA into DNA and amplification of specific DNA targets using polymerase chain reaction (PCR). It is primarily used to measure the amount of a specific RNA. This is achieved by monitoring the amplification reaction using fluorescence, a technique called real-time PCR or quantitative PCR (qPCR). Confusion can arise because some authors use the acronym RT-PCR to denote real-time PCR. In this article, RT-PCR will denote Reverse Transcription PCR. Combined RT-PCR and qPCR are routinely used for analysis of gene expression and quantification of viral RNA in research and clinical settings.
An assay is an investigative (analytic) procedure in laboratory medicine, mining, pharmacology, environmental biology and molecular biology for qualitatively assessing or quantitatively measuring the presence, amount, or functional activity of a target entity. The measured entity is often called the analyte, the measurand, or the target of the assay. The analyte can be a drug, biochemical substance, chemical element or compound, or cell in an organism or organic sample. An assay usually aims to measure an analyte's intensive property and express it in the relevant measurement unit.
The first isolation of deoxyribonucleic acid (DNA) was done in 1869 by Friedrich Miescher. DNA extraction is the process of isolating DNA from the cells of an organism isolated from a sample, typically a biological sample such as blood, saliva, or tissue. It involves breaking open the cells, removing proteins and other contaminants, and purifying the DNA so that it is free of other cellular components. The purified DNA can then be used for downstream applications such as PCR, sequencing, or cloning. Currently, it is a routine procedure in molecular biology or forensic analyses.
Capillary electrophoresis (CE) is a family of electrokinetic separation methods performed in submillimeter diameter capillaries and in micro- and nanofluidic channels. Very often, CE refers to capillary zone electrophoresis (CZE), but other electrophoretic techniques including capillary gel electrophoresis (CGE), capillary isoelectric focusing (CIEF), capillary isotachophoresis and micellar electrokinetic chromatography (MEKC) belong also to this class of methods. In CE methods, analytes migrate through electrolyte solutions under the influence of an electric field. Analytes can be separated according to ionic mobility and/or partitioning into an alternate phase via non-covalent interactions. Additionally, analytes may be concentrated or "focused" by means of gradients in conductivity and pH.

Sanger sequencing is a method of DNA sequencing that involves electrophoresis and is based on the random incorporation of chain-terminating dideoxynucleotides by DNA polymerase during in vitro DNA replication. After first being developed by Frederick Sanger and colleagues in 1977, it became the most widely used sequencing method for approximately 40 years. An automated instrument using slab gel electrophoresis and fluorescent labels was first commercialized by Applied Biosystems in March 1987. Later, automated slab gels were replaced with automated capillary array electrophoresis. More recently, higher volume Sanger sequencing has been replaced by next generation sequencing methods, especially for large-scale, automated genome analyses. However, the Sanger method remains in wide use for smaller-scale projects and for validation of deep sequencing results. It still has the advantage over short-read sequencing technologies in that it can produce DNA sequence reads of > 500 nucleotides and maintains a very low error rate with accuracies around 99.99%. Sanger sequencing is still actively being used in efforts for public health initiatives such as sequencing the spike protein from SARS-CoV-2 as well as for the surveillance of norovirus outbreaks through the Center for Disease Control and Prevention's (CDC) CaliciNet surveillance network.

An electropherogram is a record or chart produced when electrophoresis is used in an analytical technique, primarily in the fields of forensic biology, molecular biology, and biochemistry. The method plots data points that represent a specific time and fluorescence intensity at various wavelengths of light to represent a DNA profile.
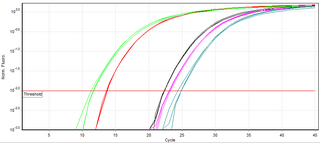
A real-time polymerase chain reaction is a laboratory technique of molecular biology based on the polymerase chain reaction (PCR). It monitors the amplification of a targeted DNA molecule during the PCR, not at its end, as in conventional PCR. Real-time PCR can be used quantitatively and semi-quantitatively.

Genetic analysis is the overall process of studying and researching in fields of science that involve genetics and molecular biology. There are a number of applications that are developed from this research, and these are also considered parts of the process. The base system of analysis revolves around general genetics. Basic studies include identification of genes and inherited disorders. This research has been conducted for centuries on both a large-scale physical observation basis and on a more microscopic scale. Genetic analysis can be used generally to describe methods both used in and resulting from the sciences of genetics and molecular biology, or to applications resulting from this research.
Terminal restriction fragment length polymorphism is a molecular biology technique for profiling of microbial communities based on the position of a restriction site closest to a labelled end of an amplified gene. The method is based on digesting a mixture of PCR amplified variants of a single gene using one or more restriction enzymes and detecting the size of each of the individual resulting terminal fragments using a DNA sequencer. The result is a graph image where the x-axis represents the sizes of the fragment and the y-axis represents their fluorescence intensity.
Multiplex ligation-dependent probe amplification (MLPA) is a variation of the multiplex polymerase chain reaction that permits amplification of multiple targets with only a single primer pair. It detects copy number changes at the molecular level, and software programs are used for analysis. Identification of deletions or duplications can indicate pathogenic mutations, thus MLPA is an important diagnostic tool used in clinical pathology laboratories worldwide.
Digital polymerase chain reaction is a biotechnological refinement of conventional polymerase chain reaction methods that can be used to directly quantify and clonally amplify nucleic acids strands including DNA, cDNA, or RNA. The key difference between dPCR and qPCR lies in the method of measuring nucleic acids amounts, with the former being a more precise method than PCR, though also more prone to error in the hands of inexperienced users. PCR carries out one reaction per single sample. dPCR also carries out a single reaction within a sample, however the sample is separated into a large number of partitions and the reaction is carried out in each partition individually. This separation allows a more reliable collection and sensitive measurement of nucleic acid amounts. The method has been demonstrated as useful for studying variations in gene sequences—such as copy number variants and point mutations.
The RNA integrity number (RIN) is an algorithm for assigning integrity values to RNA measurements.
Melting curve analysis is an assessment of the dissociation characteristics of double-stranded DNA during heating. As the temperature is raised, the double strand begins to dissociate leading to a rise in the absorbance intensity, hyperchromicity. The temperature at which 50% of DNA is denatured is known as the melting temperature. Measurement of melting temperature can help us predict species by just studying the melting temperature. This is because every organism has a specific melting curve.
Multiplex polymerase chain reaction refers to the use of polymerase chain reaction to amplify several different DNA sequences simultaneously. This process amplifies DNA in samples using multiple primers and a temperature-mediated DNA polymerase in a thermal cycler. The primer design for all primers pairs has to be optimized so that all primer pairs can work at the same annealing temperature during PCR.
Virus quantification is counting or calculating the number of virus particles (virions) in a sample to determine the virus concentration. It is used in both research and development (R&D) in academic and commercial laboratories as well as in production situations where the quantity of virus at various steps is an important variable that must be monitored. For example, the production of virus-based vaccines, recombinant proteins using viral vectors, and viral antigens all require virus quantification to continually monitor and/or modify the process in order to optimize product quality and production yields and to respond to ever changing demands and applications. Other examples of specific instances where viruses need to be quantified include clone screening, multiplicity of infection (MOI) optimization, and adaptation of methods to cell culture.
Community fingerprinting is a set of molecular biology techniques that can be used to quickly profile the diversity of a microbial community. Rather than directly identifying or counting individual cells in an environmental sample, these techniques show how many variants of a gene are present. In general, it is assumed that each different gene variant represents a different type of microbe. Community fingerprinting is used by microbiologists studying a variety of microbial systems to measure biodiversity or track changes in community structure over time. The method analyzes environmental samples by assaying genomic DNA. This approach offers an alternative to microbial culturing, which is important because most microbes cannot be cultured in the laboratory. Community fingerprinting does not result in identification of individual microbe species; instead, it presents an overall picture of a microbial community. These methods are now largely being replaced by high throughput sequencing, such as targeted microbiome analysis and metagenomics.
References
- 1 2 "Relative fluorescence unit (RFU)", DNA.gov: Glossary, April 2011, webpage: DNA-gov-RFU.
- ↑ "Thresholds", NFSTC.org, 2010, webpage: .
- ↑ Gertsch J, Güttinger M, Sticher O, Heilmann J (2002). "Relative quantification of mRNA levels in Jurkat T cells with RT-real time-PCR (RT-rt-PCR): new possibilities for the screening of anti-inflammatory and cytotoxic compounds". Pharm Res. 19 (8): 1236–1243. doi:10.1023/A:1019818814336. PMID 12240952. S2CID 17465967.
- 1 2 3 4 5 6 7 8 9 10 "Advancing Justice Through DNA Technology", National Forensic Science Technology Center (NFSTC), 2011, web: NFS2.