
A retrovirus is a type of virus that inserts a copy of its RNA genome into the DNA of a host cell that it invades, thus changing the genome of that cell. Once inside the host cell's cytoplasm, the virus uses its own reverse transcriptase enzyme to produce DNA from its RNA genome, the reverse of the usual pattern, thus retro (backwards). The new DNA is then incorporated into the host cell genome by an integrase enzyme, at which point the retroviral DNA is referred to as a provirus. The host cell then treats the viral DNA as part of its own genome, transcribing and translating the viral genes along with the cell's own genes, producing the proteins required to assemble new copies of the virus.

An RNA virus is a virus which has ribonucleic acid (RNA) as its genetic material. The nucleic acid is usually single-stranded RNA (ssRNA) but it may be double-stranded (dsRNA). Notable human diseases caused by RNA viruses include the common cold, influenza, SARS, MERS, COVID-19, Dengue Virus, hepatitis C, hepatitis E, West Nile fever, Ebola virus disease, rabies, polio, mumps, and measles.

A poliovirus, the causative agent of polio, is a serotype of the species Enterovirus C, in the family of Picornaviridae. There are three poliovirus serotypes.

Flavivirus is a genus of positive-strand RNA viruses in the family Flaviviridae. The genus includes the West Nile virus, dengue virus, tick-borne encephalitis virus, yellow fever virus, Zika virus and several other viruses which may cause encephalitis, as well as insect-specific flaviviruses (ISFs) such as cell fusing agent virus (CFAV), Palm Creek virus (PCV), and Parramatta River virus (PaRV). While dual-host flaviviruses can infect vertebrates as well as arthropods, insect-specific flaviviruses are restricted to their competent arthropods. The means by which flaviviruses establish persistent infection in their competent vectors and cause disease in humans depends upon several virus-host interactions, including the intricate interplay between flavivirus-encoded immune antagonists and the host antiviral innate immune effector molecules.

Picornaviruses are a group of related nonenveloped RNA viruses which infect vertebrates including fish, mammals, and birds. They are viruses that represent a large family of small, positive-sense, single-stranded RNA viruses with a 30 nm icosahedral capsid. The viruses in this family can cause a range of diseases including the common cold, poliomyelitis, meningitis, hepatitis, and paralysis.

Ribonuclease is a type of nuclease that catalyzes the degradation of RNA into smaller components. Ribonucleases can be divided into endoribonucleases and exoribonucleases, and comprise several sub-classes within the EC 2.7 and 3.1 classes of enzymes.

Ribonuclease H is a family of non-sequence-specific endonuclease enzymes that catalyze the cleavage of RNA in an RNA/DNA substrate via a hydrolytic mechanism. Members of the RNase H family can be found in nearly all organisms, from bacteria to archaea to eukaryotes.

An arenavirus is a bisegmented ambisense RNA virus that is a member of the family Arenaviridae. These viruses infect rodents and occasionally humans. A class of novel, highly divergent arenaviruses, properly known as reptarenaviruses, have also been discovered which infect snakes to produce inclusion body disease. At least eight arenaviruses are known to cause human disease. The diseases derived from arenaviruses range in severity. Aseptic meningitis, a severe human disease that causes inflammation covering the brain and spinal cord, can arise from the lymphocytic choriomeningitis virus. Hemorrhagic fever syndromes, including Lassa fever, are derived from infections such as Guanarito virus, Junin virus, Lassa virus, Lujo virus, Machupo virus, Sabia virus, or Whitewater Arroyo virus. Because of the epidemiological association with rodents, some arenaviruses and bunyaviruses are designated as roboviruses.
The genome and proteins of HIV have been the subject of extensive research since the discovery of the virus in 1983. "In the search for the causative agent, it was initially believed that the virus was a form of the Human T-cell leukemia virus (HTLV), which was known at the time to affect the human immune system and cause certain leukemias. However, researchers at the Pasteur Institute in Paris isolated a previously unknown and genetically distinct retrovirus in patients with AIDS which was later named HIV." Each virion comprises a viral envelope and associated matrix enclosing a capsid, which itself encloses two copies of the single-stranded RNA genome and several enzymes. The discovery of the virus itself occurred two years following the report of the first major cases of AIDS-associated illnesses.

Ribonuclease L or RNase L, known sometimes as ribonuclease 4 or 2'-5' oligoadenylate synthetase-dependent ribonuclease — is an interferon (IFN)-induced ribonuclease which, upon activation, destroys all RNA within the cell. RNase L is an enzyme that in humans is encoded by the RNASEL gene.
The murine leukemia viruses are retroviruses named for their ability to cause cancer in murine (mouse) hosts. Some MLVs may infect other vertebrates. MLVs include both exogenous and endogenous viruses. Replicating MLVs have a positive sense, single-stranded RNA (ssRNA) genome that replicates through a DNA intermediate via the process of reverse transcription.
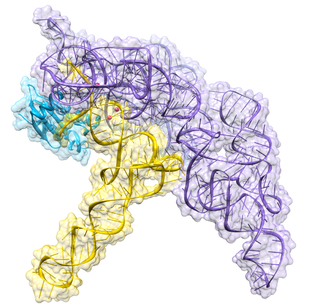
Ribonuclease P is a type of ribonuclease which cleaves RNA. RNase P is unique from other RNases in that it is a ribozyme – a ribonucleic acid that acts as a catalyst in the same way that a protein-based enzyme would. Its function is to cleave off an extra, or precursor, sequence of RNA on tRNA molecules. Further, RNase P is one of two known multiple turnover ribozymes in nature, the discovery of which earned Sidney Altman and Thomas Cech the Nobel Prize in Chemistry in 1989: in the 1970s, Altman discovered the existence of precursor tRNA with flanking sequences and was the first to characterize RNase P and its activity in processing of the 5' leader sequence of precursor tRNA. Recent findings also reveal that RNase P has a new function. It has been shown that human nuclear RNase P is required for the normal and efficient transcription of various small noncoding RNAs, such as tRNA, 5S rRNA, SRP RNA and U6 snRNA genes, which are transcribed by RNA polymerase III, one of three major nuclear RNA polymerases in human cells.
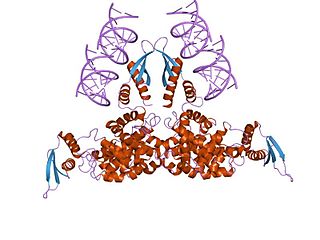
Ribonuclease III (BRENDA 3.1.26.3) is a type of ribonuclease that recognizes dsRNA and cleaves it at specific targeted locations to transform them into mature RNAs. These enzymes are a group of endoribonucleases that are characterized by their ribonuclease domain, which is labelled the RNase III domain. They are ubiquitous compounds in the cell and play a major role in pathways such as RNA precursor synthesis, RNA Silencing, and the pnp autoregulatory mechanism.
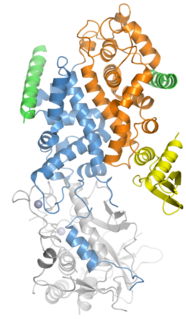
Drosha is a Class 2 ribonuclease III enzyme that in humans is encoded by the DROSHA gene. It is the primary nuclease that executes the initiation step of miRNA processing in the nucleus. It works closely with DGCR8 and in correlation with Dicer. It has been found significant in clinical knowledge for cancer prognosis and HIV-1 replication.

RNA-dependent RNA polymerase (RdRp) or RNA replicase is an enzyme that catalyzes the replication of RNA from an RNA template. Specifically, it catalyzes synthesis of the RNA strand complementary to a given RNA template. This is in contrast to typical DNA-dependent RNA polymerases, which all organisms use to catalyze the transcription of RNA from a DNA template.

The Coronavirus packaging signal is a conserved cis-regulatory element found in Betacoronavirus. It has an important role in regulating the packaging of the viral genome into the capsid. As part of the viral life cycle, within the infected cell, the viral genome becomes associated with viral proteins and assembles into new infective progeny viruses. This process is called packaging and is vital for viral replication.

The hepatitis delta virus (HDV) ribozyme is a non-coding RNA found in the hepatitis delta virus that is necessary for viral replication and is the only known human virus that utilizes ribozyme activity to infect its host. The ribozyme acts to process the RNA transcripts to unit lengths in a self-cleavage reaction during replication of the hepatitis delta virus, which is thought to propagate by a double rolling circle mechanism. The ribozyme is active in vivo in the absence of any protein factors and was the fastest known naturally occurring self-cleaving RNA at the time of its discovery.
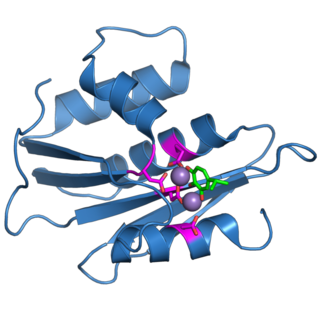
The retroviral ribonuclease H is a catalytic domain of the retroviral reverse transcriptase (RT) enzyme. The RT enzyme is used to generate complementary DNA (cDNA) from the retroviral RNA genome. This process is called reverse transcription. To complete this complex process, the retroviral RT enzymes need to adopt a multifunctional nature. They therefore possess 3 of the following biochemical activities: RNA-dependent DNA polymerase, ribonuclease H, and DNA-dependent DNA polymerase activities. Like all RNase H enzymes, the retroviral RNase H domain cleaves DNA/RNA duplexes and will not degrade DNA or unhybridized RNA.

Alphacoronaviruses (Alpha-CoV) are members of the first of the four genera of coronaviruses. They are positive-sense, single-stranded RNA viruses that infect mammals, including humans. They have spherical virions with club-shaped surface projections formed by trimers of the spike protein, and a viral envelope.

The first step of transcription for some negative, single-stranded RNA viruses is cap snatching, in which the first 10 to 20 residues of a host cell RNA are removed (snatched) and used as the 5′ cap and primer to initiate the synthesis of the nascent viral mRNA. The viral RNA-dependent RNA polymerase (RdRp) can then proceed to replicate the negative-sense genome from the positive-sense template. Cap-snatching also explains why some viral mRNA have 5’ terminal extensions of 10-20 nucleotides that are not encoded for in the genome. Examples of viruses that engage in cap-snatching include influenza viruses (Orthomyxoviridae), Lassa virus (Arenaviridae), hantaan virus (Hantaviridae) and rift valley fever virus (Phenuiviridae). Most viruses snatch 15-20 nucleotides except for the families Arenaviridae and Nairoviridae and the genus Thogotovirus (Orthomyxoviridae) which use a shorter strand.



















