Related Research Articles
Molecular biology is a branch of biology that seeks to understand the molecular basis of biological activity in and between cells, including biomolecular synthesis, modification, mechanisms, and interactions.
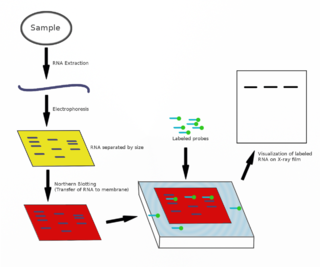
The northern blot, or RNA blot, is a technique used in molecular biology research to study gene expression by detection of RNA in a sample.
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, research, and forensics. Commonly made in the laboratory by solid-phase chemical synthesis, these small fragments of nucleic acids can be manufactured as single-stranded molecules with any user-specified sequence, and so are vital for artificial gene synthesis, polymerase chain reaction (PCR), DNA sequencing, molecular cloning and as molecular probes. In nature, oligonucleotides are usually found as small RNA molecules that function in the regulation of gene expression, or are degradation intermediates derived from the breakdown of larger nucleic acid molecules.

A DNA microarray is a collection of microscopic DNA spots attached to a solid surface. Scientists use DNA microarrays to measure the expression levels of large numbers of genes simultaneously or to genotype multiple regions of a genome. Each DNA spot contains picomoles of a specific DNA sequence, known as probes. These can be a short section of a gene or other DNA element that are used to hybridize a cDNA or cRNA sample under high-stringency conditions. Probe-target hybridization is usually detected and quantified by detection of fluorophore-, silver-, or chemiluminescence-labeled targets to determine relative abundance of nucleic acid sequences in the target. The original nucleic acid arrays were macro arrays approximately 9 cm × 12 cm and the first computerized image based analysis was published in 1981. It was invented by Patrick O. Brown. An example of its application is in SNPs arrays for polymorphisms in cardiovascular diseases, cancer, pathogens and GWAS analysis. It is also used for the identification of structural variations and the measurement of gene expression.

In molecular biology and biotechnology, a fluorescent tag, also known as a fluorescent label or fluorescent probe, is a molecule that is attached chemically to aid in the detection of a biomolecule such as a protein, antibody, or amino acid. Generally, fluorescent tagging, or labeling, uses a reactive derivative of a fluorescent molecule known as a fluorophore. The fluorophore selectively binds to a specific region or functional group on the target molecule and can be attached chemically or biologically. Various labeling techniques such as enzymatic labeling, protein labeling, and genetic labeling are widely utilized. Ethidium bromide, fluorescein and green fluorescent protein are common tags. The most commonly labelled molecules are antibodies, proteins, amino acids and peptides which are then used as specific probes for detection of a particular target.
Comparative genomic hybridization (CGH) is a molecular cytogenetic method for analysing copy number variations (CNVs) relative to ploidy level in the DNA of a test sample compared to a reference sample, without the need for culturing cells. The aim of this technique is to quickly and efficiently compare two genomic DNA samples arising from two sources, which are most often closely related, because it is suspected that they contain differences in terms of either gains or losses of either whole chromosomes or subchromosomal regions. This technique was originally developed for the evaluation of the differences between the chromosomal complements of solid tumor and normal tissue, and has an improved resolution of 5–10 megabases compared to the more traditional cytogenetic analysis techniques of giemsa banding and fluorescence in situ hybridization (FISH) which are limited by the resolution of the microscope utilized.
In molecular biology, a hybridization probe (HP) is a fragment of DNA or RNA, usually 15–10000 nucleotides long, which can be radioactively or fluorescently labeled. HPs can be used to detect the presence of nucleotide sequences in analyzed RNA or DNA that are complementary to the sequence in the probe. The labeled probe is first denatured into single stranded DNA (ssDNA) and then hybridized to the target ssDNA or RNA immobilized on a membrane or in situ.

Fluorescence in situ hybridization (FISH) is a molecular cytogenetic technique that uses fluorescent probes that bind to only particular parts of a nucleic acid sequence with a high degree of sequence complementarity. It was developed by biomedical researchers in the early 1980s to detect and localize the presence or absence of specific DNA sequences on chromosomes. Fluorescence microscopy can be used to find out where the fluorescent probe is bound to the chromosomes. FISH is often used for finding specific features in DNA for use in genetic counseling, medicine, and species identification. FISH can also be used to detect and localize specific RNA targets in cells, circulating tumor cells, and tissue samples. In this context, it can help define the spatial-temporal patterns of gene expression within cells and tissues.
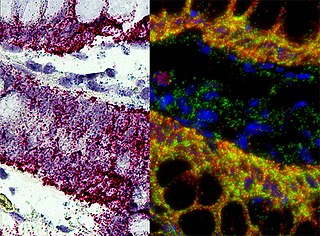
In situ hybridization (ISH) is a type of hybridization that uses a labeled complementary DNA, RNA or modified nucleic acid strand to localize a specific DNA or RNA sequence in a portion or section of tissue or if the tissue is small enough, in the entire tissue, in cells, and in circulating tumor cells (CTCs). This is distinct from immunohistochemistry, which usually localizes proteins in tissue sections.
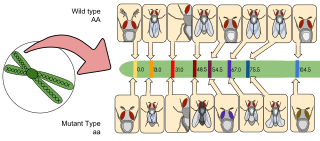
Gene mapping or genome mapping describes the methods used to identify the location of a gene on a chromosome and the distances between genes. Gene mapping can also describe the distances between different sites within a gene.
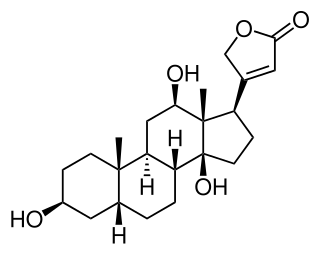
Digoxigenin (DIG) is a steroid found exclusively in the flowers and leaves of the plants Digitalis purpurea, Digitalis orientalis and Digitalis lanata (foxgloves), where it is attached to sugars, to form the glycosides.
In molecular biology and genetics, the sense of a nucleic acid molecule, particularly of a strand of DNA or RNA, refers to the nature of the roles of the strand and its complement in specifying a sequence of amino acids. Depending on the context, sense may have slightly different meanings. For example, the negative-sense strand of DNA is equivalent to the template strand, whereas the positive-sense strand is the non-template strand whose nucleotide sequence is equivalent to the sequence of the mRNA transcript.

Molecular cytogenetics combines two disciplines, molecular biology and cytogenetics, and involves the analysis of chromosome structure to help distinguish normal and cancer-causing cells. Human cytogenetics began in 1956 when it was discovered that normal human cells contain 46 chromosomes. However, the first microscopic observations of chromosomes were reported by Arnold, Flemming, and Hansemann in the late 1800s. Their work was ignored for decades until the actual chromosome number in humans was discovered as 46. In 1879, Arnold examined sarcoma and carcinoma cells having very large nuclei. Today, the study of molecular cytogenetics can be useful in diagnosing and treating various malignancies such as hematological malignancies, brain tumors, and other precursors of cancer. The field is overall focused on studying the evolution of chromosomes, more specifically the number, structure, function, and origin of chromosome abnormalities. It includes a series of techniques referred to as fluorescence in situ hybridization, or FISH, in which DNA probes are labeled with different colored fluorescent tags to visualize one or more specific regions of the genome. Introduced in the 1980s, FISH uses probes with complementary base sequences to locate the presence or absence of the specific DNA regions. FISH can either be performed as a direct approach to metaphase chromosomes or interphase nuclei. Alternatively, an indirect approach can be taken in which the entire genome can be assessed for copy number changes using virtual karyotyping. Virtual karyotypes are generated from arrays made of thousands to millions of probes, and computational tools are used to recreate the genome in silico.

A nucleic acid test (NAT) is a technique used to detect a particular nucleic acid sequence and thus usually to detect and identify a particular species or subspecies of organism, often a virus or bacterium that acts as a pathogen in blood, tissue, urine, etc. NATs differ from other tests in that they detect genetic materials rather than antigens or antibodies. Detection of genetic materials allows an early diagnosis of a disease because the detection of antigens and/or antibodies requires time for them to start appearing in the bloodstream. Since the amount of a certain genetic material is usually very small, many NATs include a step that amplifies the genetic material—that is, makes many copies of it. Such NATs are called nucleic acid amplification tests (NAATs). There are several ways of amplification, including polymerase chain reaction (PCR), strand displacement assay (SDA), transcription mediated assay (TMA), and loop-mediated isothermal amplification (LAMP).
Flow-FISH is a cytogenetic technique to quantify the copy number of RNA or specific repetitive elements in genomic DNA of whole cell populations via the combination of flow cytometry with cytogenetic fluorescent in situ hybridization staining protocols.
Chromogenic in situ hybridization (CISH) is a cytogenetic technique that combines the chromogenic signal detection method of immunohistochemistry (IHC) techniques with in situ hybridization. It was developed around the year 2000 as an alternative to fluorescence in situ hybridization (FISH) for detection of HER-2/neu oncogene amplification. CISH is similar to FISH in that they are both in situ hybridization techniques used to detect the presence or absence of specific regions of DNA. However, CISH is much more practical in diagnostic laboratories because it uses bright-field microscopes rather than the more expensive and complicated fluorescence microscopes used in FISH.
Quantitative Fluorescent in situ hybridization (Q-FISH) is a cytogenetic technique based on the traditional FISH methodology. In Q-FISH, the technique uses labelled synthetic DNA mimics called peptide nucleic acid (PNA) oligonucleotides to quantify target sequences in chromosomal DNA using fluorescent microscopy and analysis software. Q-FISH is most commonly used to study telomere length, which in vertebrates are repetitive hexameric sequences (TTAGGG) located at the distal end of chromosomes. Telomeres are necessary at chromosome ends to prevent DNA-damage responses as well as genome instability. To this day, the Q-FISH method continues to be utilized in the field of telomere research.
In molecular biology, hybridization is a phenomenon in which single-stranded deoxyribonucleic acid (DNA) or ribonucleic acid (RNA) molecules anneal to complementary DNA or RNA. Though a double-stranded DNA sequence is generally stable under physiological conditions, changing these conditions in the laboratory will cause the molecules to separate into single strands. These strands are complementary to each other but may also be complementary to other sequences present in their surroundings. Lowering the surrounding temperature allows the single-stranded molecules to anneal or “hybridize” to each other.
A hybridization assay comprises any form of quantifiable hybridization i.e. the quantitative annealing of two complementary strands of nucleic acids, known as nucleic acid hybridization.
Spatial transcriptomics is a method for assigning cell types to their locations in the histological sections. It comprises an important part of spatial biology. Recent work demonstrated that the subcellular localization of mRNA molecules, for example, in the nucleus can also be studied.
References
- ↑ Lackie, John (2010). A Dictionary of Biomedicine. Oxford University Press. ISBN 9780199549351.
- ↑ Watson, J.D.; F.H.C., Crick (1953). "Molecular structure of nucleic acids: A structure for deoxyribose nucleic acid". Nature. 171 (4356): 737–738. doi:10.1038/171737a0. PMID 13054692. S2CID 4253007.
- ↑ Gall, J.G.; Pardue, M.L. (1969). "Formation and detection of RNA-DNA hybrid molecules in cytological preparations". Proceedings of the National Academy of Sciences. 63 (2): 378–383. doi: 10.1073/pnas.63.2.378 . PMC 223575 . PMID 4895535.
- ↑ Rudkin, G.T.; Stollar, B.D. (1977). "High resolution of DNA-RNA hybrids in situ by indirect immunofluorescence". Nature. 265 (5593): 472–474. doi:10.1038/265472a0. PMID 401954. S2CID 4165112.
- ↑ O'Connor, Clare (2008). "Fluorescence In Situ Hybridization (FISH)". Nature Education. 1: 171.
- ↑ Katona, István; Urbán, Gabriella M.; Wallace, Matthew; Ledent, Catherine; Jung, Kwang-Mook; Piomelli, Daniele; Mackie, Ken; Freund, Tamás F. (2006-05-24). "Molecular Composition of the Endocannabinoid System at Glutamatergic Synapses". Journal of Neuroscience. 26 (21): 5628–5637. doi:10.1523/JNEUROSCI.0309-06.2006. ISSN 0270-6474. PMID 16723519.
- ↑ Lajtha, Abel (2007). Handbook of Neurochemistry and Molecular Neurobiology Practical Neurochemistry Method. United States: Springer ScienceþBusiness Media. p. 364. ISBN 9780387303598.
- ↑ Clark, Melody (1996). In Situ Hybridization. ISBN 9783527308859.
- ↑ D.R., Kapczy (2001). "Detection of In Ovo-Inoculated Infectious Bronchitis Virus by Immunohistochemistry and In Situ Hybridization with a Riboprobe in Epithelial Cells of the Lung and Cloaca". Avian Diseases. 46 (3 (Jul.– Sep., 2002)): 679–685. doi:10.1637/0005-2086(2002)046[0679:doioii]2.0.co;2. S2CID 12144776.
- ↑ Speicher, M.R.; et al. (2005). "The new cytogenetics: blurring the boundaries with molecular biology". Nature Reviews Genetics. 6 (10): 782–92. doi:10.1038/nrg1692. PMID 16145555. S2CID 15023775.
- ↑ McNeil, N.; Ried, T. (2000). "Novel molecular cytogenetic techniques for identifying complex chromosomal rearrangements: technology and applications in molecular medicine". Expert Reviews in Molecular Medicine. 2000: 1–14. doi:10.1017/S1462399400001940. PMID 14585138. S2CID 196593251.
Baynes, John W.; Marek H. Dominiczak (2005). Medical Biochemistry 2nd. Edition . Elsevier Mosby. p. 477. ISBN 978-0-7234-3341-5.