
The alpha helix (α-helix) is a common motif in the secondary structure of proteins and is a right hand-helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid located four residues earlier along the protein sequence.

Deoxyribonucleic acid is a molecule composed of two polynucleotide chains that coil around each other to form a double helix carrying genetic instructions for the development, functioning, growth and reproduction of all known organisms and many viruses. DNA and ribonucleic acid (RNA) are nucleic acids. Alongside proteins, lipids and complex carbohydrates (polysaccharides), nucleic acids are one of the four major types of macromolecules that are essential for all known forms of life.

Ribonucleic acid (RNA) is a polymeric molecule essential in various biological roles in coding, decoding, regulation and expression of genes. RNA and deoxyribonucleic acid (DNA) are nucleic acids. Along with lipids, proteins, and carbohydrates, nucleic acids constitute one of the four major macromolecules essential for all known forms of life. Like DNA, RNA is assembled as a chain of nucleotides, but unlike DNA, RNA is found in nature as a single strand folded onto itself, rather than a paired double strand. Cellular organisms use messenger RNA (mRNA) to convey genetic information that directs synthesis of specific proteins. Many viruses encode their genetic information using an RNA genome.

A spliceosome is a large ribonucleoprotein (RNP) complex found primarily within the nucleus of eukaryotic cells. The spliceosome is assembled from small nuclear RNAs (snRNA) and numerous proteins (Small nuclear RNA molecules bind to specific proteins to form a small nuclear ribonucleoprotein complex, which in turn combines with other snRNPs to form a large ribonucleoprotein complex called a spliceosome). The spliceosome removes introns from a transcribed pre-mRNA, a type of primary transcript. This process is generally referred to as splicing. An analogy is a film editor, who selectively cuts out irrelevant or incorrect material from the initial film and sends the cleaned-up version to the director for the final cut.
In a chain-like biological molecule, such as a protein or nucleic acid, a structural motif is a common three-dimensional structure that appears in a variety of different, evolutionarily unrelated molecules. A structural motif does not have to be associated with a sequence motif; it can be represented by different and completely unrelated sequences in different proteins or RNA.

RecA is a 38 kilodalton protein essential for the repair and maintenance of DNA. A RecA structural and functional homolog has been found in every species in which one has been seriously sought and serves as an archetype for this class of homologous DNA repair proteins. The homologous protein is called RAD51 in eukaryotes and RadA in archaea.

In molecular biology, the term double helix refers to the structure formed by double-stranded molecules of nucleic acids such as DNA. The double helical structure of a nucleic acid complex arises as a consequence of its secondary structure, and is a fundamental component in determining its tertiary structure. The term entered popular culture with the publication in 1968 of The Double Helix: A Personal Account of the Discovery of the Structure of DNA by James Watson.
Extrachromosomal DNA is any DNA that is found off the chromosomes, either inside or outside the nucleus of a cell. Most DNA in an individual genome is found in chromosomes contained in the nucleus. Multiple forms of extrachromosomal DNA exist and serve important biological functions, e.g. they can play a role in disease, such as ecDNA in cancer.

Biomolecular structure is the intricate folded, three-dimensional shape that is formed by a molecule of protein, DNA, or RNA, and that is important to its function. The structure of these molecules may be considered at any of several length scales ranging from the level of individual atoms to the relationships among entire protein subunits. This useful distinction among scales is often expressed as a decomposition of molecular structure into four levels: primary, secondary, tertiary, and quaternary. The scaffold for this multiscale organization of the molecule arises at the secondary level, where the fundamental structural elements are the molecule's various hydrogen bonds. This leads to several recognizable domains of protein structure and nucleic acid structure, including such secondary-structure features as alpha helixes and beta sheets for proteins, and hairpin loops, bulges, and internal loops for nucleic acids. The terms primary, secondary, tertiary, and quaternary structure were introduced by Kaj Ulrik Linderstrøm-Lang in his 1951 Lane Medical Lectures at Stanford University.
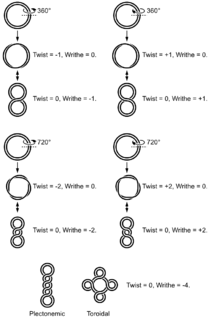
DNA supercoiling refers to the over- or under-winding of a DNA strand, and is an expression of the strain on that strand. Supercoiling is important in a number of biological processes, such as compacting DNA, and by regulating access to the genetic code, DNA supercoiling strongly affects DNA metabolism and possibly gene expression. Additionally, certain enzymes such as topoisomerases are able to change DNA topology to facilitate functions such as DNA replication or transcription. Mathematical expressions are used to describe supercoiling by comparing different coiled states to relaxed B-form DNA.
A helix bundle is a small protein fold composed of several alpha helices that are usually nearly parallel or antiparallel to each other.

In molecular biology, LSm proteins are a family of RNA-binding proteins found in virtually every cellular organism. LSm is a contraction of 'like Sm', because the first identified members of the LSm protein family were the Sm proteins. LSm proteins are defined by a characteristic three-dimensional structure and their assembly into rings of six or seven individual LSm protein molecules, and play a large number of various roles in mRNA processing and regulation.

The 5S ribosomal RNA is an approximately 120 nucleotide-long ribosomal RNA molecule with a mass of 40 kDa. It is a structural and functional component of the large subunit of the ribosome in all domains of life, with the exception of mitochondrial ribosomes of fungi and animals. The designation 5S refers to the molecule's sedimentation velocity in an ultracentrifuge, which is measured in Svedberg units (S).

The signal recognition particle RNA, is part of the signal recognition particle (SRP) ribonucleoprotein complex. SRP recognizes the signal peptide and binds to the ribosome, halting protein synthesis. SRP-receptor is a protein that is embedded in a membrane, and which contains a transmembrane pore. When the SRP-ribosome complex binds to SRP-receptor, SRP releases the ribosome and drifts away. The ribosome resumes protein synthesis, but now the protein is moving through the SRP-receptor transmembrane pore.

Molecular models of DNA structures are representations of the molecular geometry and topology of deoxyribonucleic acid (DNA) molecules using one of several means, with the aim of simplifying and presenting the essential, physical and chemical, properties of DNA molecular structures either in vivo or in vitro. These representations include closely packed spheres made of plastic, metal wires for skeletal models, graphic computations and animations by computers, artistic rendering. Computer molecular models also allow animations and molecular dynamics simulations that are very important for understanding how DNA functions in vivo.

Nucleic acid tertiary structure is the three-dimensional shape of a nucleic acid polymer. RNA and DNA molecules are capable of diverse functions ranging from molecular recognition to catalysis. Such functions require a precise three-dimensional tertiary structure. While such structures are diverse and seemingly complex, they are composed of recurring, easily recognizable tertiary structure motifs that serve as molecular building blocks. Some of the most common motifs for RNA and DNA tertiary structure are described below, but this information is based on a limited number of solved structures. Many more tertiary structural motifs will be revealed as new RNA and DNA molecules are structurally characterized.

Nucleic acid structure refers to the structure of nucleic acids such as DNA and RNA. Chemically speaking, DNA and RNA are very similar. Nucleic acid structure is often divided into four different levels: primary, secondary, tertiary, and quaternary.

Nucleic acid secondary structure is the basepairing interactions within a single nucleic acid polymer or between two polymers. It can be represented as a list of bases which are paired in a nucleic acid molecule. The secondary structures of biological DNA's and RNA's tend to be different: biological DNA mostly exists as fully base paired double helices, while biological RNA is single stranded and often forms complex and intricate base-pairing interactions due to its increased ability to form hydrogen bonds stemming from the extra hydroxyl group in the ribose sugar.
ColE1 is a plasmid found in bacteria. Its name derives from the fact that it carries a gene for colicin E1. It also codes for immunity from this product with the imm gene. In addition, the plasmid has a series of mobility (mob) genes. Its size and the presence of a single EcoRI recognition site caused it to be considered as a vector candidate.
In cellular biology, the plasmid copy number is the number of copies of a given plasmid in a cell. To ensure survival and thus the continued propagation of the plasmid, they must regulate their copy number. If a plasmid has too high of a copy number, they may excessively burden their host by occupying too much cellular machinery and using too much energy. On the other hand, too low of a copy number may result in the plasmid not being present in all of their host's progeny. Plasmids may be either high copy number plasmids or low copy number plasmids; the regulation mechanisms between these two types are often significantly different. Biotechnology applications may involve engineering plasmids to allow a very high copy number. For example, pBR322 is a low copy number plasmid from which several very high copy number cloning vectors have been derived.















