Related Research Articles

In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, mitosis, or meiosis or other types of damage to DNA, which then may undergo error-prone repair, cause an error during other forms of repair, or cause an error during replication. Mutations may also result from substitution,insertion or deletion of segments of DNA due to mobile genetic elements.

Arabidopsis thaliana, the thale cress, mouse-ear cress or arabidopsis, is a small plant from the mustard family (Brassicaceae), native to Eurasia and Africa. Commonly found along the shoulders of roads and in disturbed land, it is generally considered a weed.

Genetic recombination is the exchange of genetic material between different organisms which leads to production of offspring with combinations of traits that differ from those found in either parent. In eukaryotes, genetic recombination during meiosis can lead to a novel set of genetic information that can be further passed on from parents to offspring. Most recombination occurs naturally and can be classified into two types: (1) interchromosomal recombination, occurring through independent assortment of alleles whose loci are on different but homologous chromosomes ; & (2) intrachromosomal recombination, occurring through crossing over.

Mosaicism or genetic mosaicism is a condition in which a multicellular organism possesses more than one genetic line as the result of genetic mutation. This means that various genetic lines resulted from a single fertilized egg. Mosaicism is one of several possible causes of chimerism, wherein a single organism is composed of cells with more than one distinct genotype.
RecQ helicase is a family of helicase enzymes initially found in Escherichia coli that has been shown to be important in genome maintenance. They function through catalyzing the reaction ATP + H2O → ADP + P and thus driving the unwinding of paired DNA and translocating in the 3' to 5' direction. These enzymes can also drive the reaction NTP + H2O → NDP + P to drive the unwinding of either DNA or RNA.
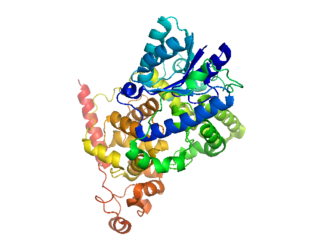
Cryptochromes are a class of flavoproteins found in plants and animals that are sensitive to blue light. They are involved in the circadian rhythms and the sensing of magnetic fields in a number of species. The name cryptochrome was proposed as a portmanteau combining the chromatic nature of the photoreceptor, and the cryptogamic organisms on which many blue-light studies were carried out.

Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids.
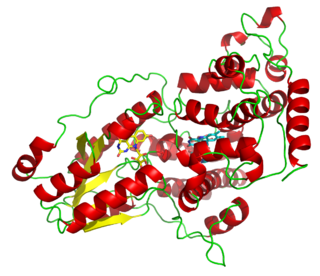
Photolyases are DNA repair enzymes that repair damage caused by exposure to ultraviolet light. These enzymes require visible light both for their own activation and for the actual DNA repair. The DNA repair mechanism involving photolyases is called photoreactivation. They mainly convert pyrimidine dimers into a normal pair of pyrimidine bases. Photo reactivation, the first DNA repair mechanism to be discovered, was described initially by Albert Kelner in 1949 and independently by Renato Dulbecco also in 1949.
Mitotic recombination is a type of genetic recombination that may occur in somatic cells during their preparation for mitosis in both sexual and asexual organisms. In asexual organisms, the study of mitotic recombination is one way to understand genetic linkage because it is the only source of recombination within an individual. Additionally, mitotic recombination can result in the expression of recessive alleles in an otherwise heterozygous individual. This expression has important implications for the study of tumorigenesis and lethal recessive alleles. Mitotic homologous recombination occurs mainly between sister chromatids subsequent to replication. Inter-sister homologous recombination is ordinarily genetically silent. During mitosis the incidence of recombination between non-sister homologous chromatids is only about 1% of that between sister chromatids.

Ku is a dimeric protein complex that binds to DNA double-strand break ends and is required for the non-homologous end joining (NHEJ) pathway of DNA repair. Ku is evolutionarily conserved from bacteria to humans. The ancestral bacterial Ku is a homodimer. Eukaryotic Ku is a heterodimer of two polypeptides, Ku70 (XRCC6) and Ku80 (XRCC5), so named because the molecular weight of the human Ku proteins is around 70 kDa and 80 kDa. The two Ku subunits form a basket-shaped structure that threads onto the DNA end. Once bound, Ku can slide down the DNA strand, allowing more Ku molecules to thread onto the end. In higher eukaryotes, Ku forms a complex with the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) to form the full DNA-dependent protein kinase, DNA-PK. Ku is thought to function as a molecular scaffold to which other proteins involved in NHEJ can bind, orienting the double-strand break for ligation.

Physcomitrella patens is a synonym of Physcomitrium patens, the spreading earthmoss. It is a moss, a bryophyte used as a model organism for studies on plant evolution, development, and physiology.

Serine/threonine-protein kinase ATR, also known as ataxia telangiectasia and Rad3-related protein (ATR) or FRAP-related protein 1 (FRP1), is an enzyme that, in humans, is encoded by the ATR gene. It is a large kinase of about 301.66 kDa. ATR belongs to the phosphatidylinositol 3-kinase-related kinase protein family. ATR is activated in response to single strand breaks, and works with ATM to ensure genome integrity.
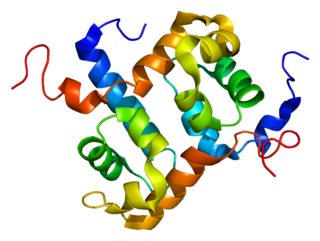
ERCC4 is a protein designated as DNA repair endonuclease XPF that in humans is encoded by the ERCC4 gene. Together with ERCC1, ERCC4 forms the ERCC1-XPF enzyme complex that participates in DNA repair and DNA recombination.

Structural maintenance of chromosomes protein 6 is a protein that in humans is encoded by the SMC6 gene.
Genome instability refers to a high frequency of mutations within the genome of a cellular lineage. These mutations can include changes in nucleic acid sequences, chromosomal rearrangements or aneuploidy. Genome instability does occur in bacteria. In multicellular organisms genome instability is central to carcinogenesis, and in humans it is also a factor in some neurodegenerative diseases such as amyotrophic lateral sclerosis or the neuromuscular disease myotonic dystrophy.
LUX or Phytoclock1 (PCL1) is a gene that codes for LUX ARRHYTHMO, a protein necessary for circadian rhythms in Arabidopsis thaliana. LUX protein associates with Early Flowering 3 (ELF3) and Early Flowering 4 (ELF4) to form the Evening Complex (EC), a core component of the Arabidopsis repressilator model of the plant circadian clock. The LUX protein functions as a transcription factor that negatively regulates Pseudo-Response Regulator 9 (PRR9), a core gene of the Midday Complex, another component of the Arabidopsis repressilator model. LUX is also associated with circadian control of hypocotyl growth factor genes PHYTOCHROME INTERACTING FACTOR 4 (PIF4) and PHYTOCHROME INTERACTING FACTOR 5 (PIF5).
Robert L. Last is a plant biochemical genomicist who studies metabolic processes that protect plants from the environment and produce products important for animal and human nutrition. His research has covered (1) production and breakdown of essential amino acids, (2) the synthesis and protective roles of Vitamin C and Vitamin E (tocopherols) as well as identification of mechanisms that protect photosystem II from damage, and (3) synthesis and biological functions of plant protective specialized metabolites. Four central questions are: (i) how are leaf and seed amino acids levels regulated, (ii.) what mechanisms protect and repair photosystem II from stress-induced damage, (iii.) how do plants produce protective metabolites in their glandular secreting trichomes (iv.) and what are the evolutionary mechanisms that contribute to the tremendous diversity of specialized metabolites that protect plants from insects and pathogens and are used as therapeutic agents.
Mutational signatures are characteristic combinations of mutation types arising from specific mutagenesis processes such as DNA replication infidelity, exogenous and endogenous genotoxin exposures, defective DNA repair pathways, and DNA enzymatic editing.
Transgenerational epigenetic inheritance in plants involves mechanisms for the passing of epigenetic marks from parent to offspring that differ from those reported in animals. There are several kinds of epigenetic markers, but they all provide a mechanism to facilitate greater phenotypic plasticity by influencing the expression of genes without altering the DNA code. These modifications represent responses to environmental input and are reversible changes to gene expression patterns that can be passed down through generations. In plants, transgenerational epigenetic inheritance could potentially represent an evolutionary adaptation for sessile organisms to quickly adapt to their changing environment.

Pal Maliga is a plant molecular biologist. He is Distinguished Professor of Plant Biology and Laboratory Director at the Waksman Institute of Microbiology, Rutgers University. He is known for developing the technology of chloroplast genome engineering in land plants and its applications in basic science and biotechnology.
References
- ↑ Gellert M (1992). "Molecular analysis of V(D)J recombination". Annu Rev Genet. 26: 425–46. doi:10.1146/annurev.ge.26.120192.002233. PMID 1482120.
- ↑ Hein K, Lorenz MG, Siebenkotten G, et al. (1998). "Processing of switch transcripts is required for targeting of antibody class switch recombination". J Exp Med. 188 (12): 2369–74. doi:10.1084/jem.188.12.2369. PMC 2212419 . PMID 9858523.
- ↑ Ramel C, Cederberg H, Magnusson J, et al. (1996). "Somatic recombination, gene amplification and cancer". Mutat Res. 353 (1–2): 85–107. Bibcode:1996MRFMM.353...85R. doi:10.1016/0027-5107(95)00243-x. PMID 8692194.
- 1 2 Lee MH, Siddoway B, Kaeser GE, Segota I, Rivera R, Romanow WJ, Liu CS, Park C, Kennedy G, Long T, Chun J (November 2018). "Somatic APP gene recombination in Alzheimer's disease and normal neurons". Nature. 563 (7733): 639–645. Bibcode:2018Natur.563..639L. doi:10.1038/s41586-018-0718-6. PMC 6391999 . PMID 30464338.
- 1 2 Swoboda P, Gal S, Hohn B, Puchta H (January 1994). "Intrachromosomal homologous recombination in whole plants". EMBO J. 13 (2): 484–9. doi:10.1002/j.1460-2075.1994.tb06283.x. PMC 394832 . PMID 8313893.
- 1 2 Masson JE, Paszkowski J (October 1997). "Arabidopsis thaliana mutants altered in homologous recombination". Proc Natl Acad Sci U S A. 94 (21): 11731–5. Bibcode:1997PNAS...9411731M. doi: 10.1073/pnas.94.21.11731 . PMC 23619 . PMID 9326679.
- 1 2 Ries G, Buchholz G, Frohnmeyer H, Hohn B (November 2000). "UV-damage-mediated induction of homologous recombination in Arabidopsis is dependent on photosynthetically active radiation". Proc Natl Acad Sci U S A. 97 (24): 13425–9. Bibcode:2000PNAS...9713425R. doi: 10.1073/pnas.230251897 . PMC 27240 . PMID 11069284.