
Förster resonance energy transfer (FRET), fluorescence resonance energy transfer, resonance energy transfer (RET) or electronic energy transfer (EET) is a mechanism describing energy transfer between two light-sensitive molecules (chromophores). A donor chromophore, initially in its electronic excited state, may transfer energy to an acceptor chromophore through nonradiative dipole–dipole coupling. The efficiency of this energy transfer is inversely proportional to the sixth power of the distance between donor and acceptor, making FRET extremely sensitive to small changes in distance.
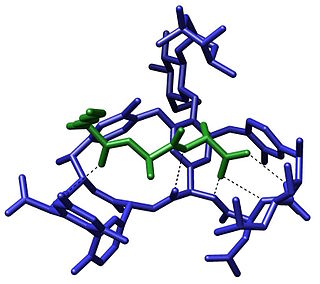
The term molecular recognition refers to the specific interaction between two or more molecules through noncovalent bonding such as hydrogen bonding, metal coordination, hydrophobic forces, van der Waals forces, π-π interactions, halogen bonding, or resonant interaction effects. In addition to these direct interactions, solvents can play a dominant indirect role in driving molecular recognition in solution. The host and guest involved in molecular recognition exhibit molecular complementarity. Exceptions are molecular containers, including e.g. nanotubes, in which portals essentially control selectivity.
Virus-like particles (VLPs) are molecules that closely resemble viruses, but are non-infectious because they contain no viral genetic material. They can be naturally occurring or synthesized through the individual expression of viral structural proteins, which can then self assemble into the virus-like structure. Combinations of structural capsid proteins from different viruses can be used to create recombinant VLPs. Both in-vivo assembly and in-vitro assembly have been successfully shown to form virus-like particles. VLPs derived from the Hepatitis B virus (HBV) and composed of the small HBV derived surface antigen (HBsAg) were described in 1968 from patient sera. VLPs have been produced from components of a wide variety of virus families including Parvoviridae, Retroviridae, Flaviviridae, Paramyxoviridae and bacteriophages. VLPs can be produced in multiple cell culture systems including bacteria, mammalian cell lines, insect cell lines, yeast and plant cells.

A catalytic triad is a set of three coordinated amino acids that can be found in the active site of some enzymes. Catalytic triads are most commonly found in hydrolase and transferase enzymes. An acid-base-nucleophile triad is a common motif for generating a nucleophilic residue for covalent catalysis. The residues form a charge-relay network to polarise and activate the nucleophile, which attacks the substrate, forming a covalent intermediate which is then hydrolysed to release the product and regenerate free enzyme. The nucleophile is most commonly a serine or cysteine amino acid, but occasionally threonine or even selenocysteine. The 3D structure of the enzyme brings together the triad residues in a precise orientation, even though they may be far apart in the sequence.

In the context of chemistry and molecular modelling, a force field is a computational method that is used to estimate the forces between atoms within molecules and also between molecules. More precisely, the force field refers to the functional form and parameter sets used to calculate the potential energy of a system of atoms or coarse-grained particles in molecular mechanics, molecular dynamics, or Monte Carlo simulations. The parameters for a chosen energy function may be derived from experiments in physics and chemistry, calculations in quantum mechanics, or both. Force fields are interatomic potentials and utilize the same concept as force fields in classical physics, with the difference that the force field parameters in chemistry describe the energy landscape, from which the acting forces on every particle are derived as a gradient of the potential energy with respect to the particle coordinates.

The Hofmeister series or lyotropic series is a classification of ions in order of their lyotrophic properties, which is the ability to salt out or salt in proteins. The effects of these changes were first worked out by Franz Hofmeister, who studied the effects of cations and anions on the solubility of proteins.

Rolipram is a selective phosphodiesterase-4 inhibitor discovered and developed by Schering AG as a potential antidepressant drug in the early 1990s. It served as a prototype molecule for several companies' drug discovery and development efforts. Rolipram was discontinued after clinical trials showed that its therapeutic window was too narrow; it could not be dosed at high enough levels to be effective without causing significant gastrointestinal side effects.

Serine hydroxymethyltransferase (SHMT) is a pyridoxal phosphate (PLP) (Vitamin B6) dependent enzyme (EC 2.1.2.1) which plays an important role in cellular one-carbon pathways by catalyzing the reversible, simultaneous conversions of L-serine to glycine and tetrahydrofolate (THF) to 5,10-Methylenetetrahydrofolate (5,10-CH2-THF). This reaction provides the largest part of the one-carbon units available to the cell.
A tandem mass tag (TMT) is a chemical label that facilitates sample multiplexing in mass spectrometry (MS)-based quantification and identification of biological macromolecules such as proteins, peptides and nucleic acids. TMT belongs to a family of reagents referred to as isobaric mass tags which are a set of molecules with the same mass, but yield reporter ions of differing mass after fragmentation. The relative ratio of the measured reporter ions represents the relative abundance of the tagged molecule, although ion suppression has a detrimental effect on accuracy. Despite these complications, TMT-based proteomics has been shown to afford higher precision than Label-free quantification. In addition to aiding in protein quantification, TMT tags can also increase the detection sensitivity of certain highly hydrophilic analytes, such as phosphopeptides, in RPLC-MS analyses.
In enzymology, carbon monoxide dehydrogenase (CODH) (EC 1.2.7.4) is an enzyme that catalyzes the chemical reaction

The adenosine A2B receptor, also known as ADORA2B, is a G-protein coupled adenosine receptor, and also denotes the human adenosine A2b receptor gene which encodes it.

Melanocortin 3 receptor (MC3R) is a protein that in humans is encoded by the MC3R gene.

Metabotropic glutamate receptor 3 (mGluR3) is an inhibitory Gi/G0-coupled G-protein coupled receptor (GPCR) generally localized to presynaptic sites of neurons in classical circuits. However, in higher cortical circuits in primates, mGluR3 are localized post-synaptically, where they strengthen rather than weaken synaptic connectivity. In humans, mGluR3 is encoded by the GRM3 gene. Deficits in mGluR3 signaling have been linked to impaired cognition in humans, and to increased risk of schizophrenia, consistent with their expanding role in cortical evolution.
Reflectins are a family of intrinsically disordered proteins evolved by a certain number of cephalopods including Euprymna scolopes and Doryteuthis opalescens to produce iridescent camouflage and signaling. The recently identified protein family is enriched in aromatic and sulfur-containing amino acids, and is utilized by certain cephalopods to refract incident light in their environment. The reflectin protein is responsible for dynamic pigmentation and iridescence in organisms. This process is “dynamic” due to its reversible properties, allowing reflectin to change an organism’s appearance in response to external factors such as needing to camouflage or send warning signals.
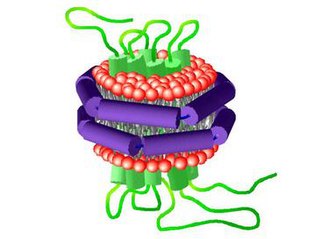
A nanodisc is a synthetic model membrane system which assists in the study of membrane proteins. Nanodiscs are discoidal proteins in which a lipid bilayer is surrounded by molecules that are amphipathic molecules including proteins, peptides, and synthetic polymers. It is composed of a lipid bilayer of phospholipids with the hydrophobic edge screened by two amphipathic proteins. These proteins are called membrane scaffolding proteins (MSP) and align in double belt formation. Nanodiscs are structurally very similar to discoidal high-density lipoproteins (HDL) and the MSPs are modified versions of apolipoprotein A1 (apoA1), the main constituent in HDL. Nanodiscs are useful in the study of membrane proteins because they can solubilise and stabilise membrane proteins and represent a more native environment than liposomes, detergent micelles, bicelles and amphipols.

E1 is one of two subunits of the envelope glycoprotein found in the hepatitis C virus. The other subunit is E2. This protein is a type 1 transmembrane protein with a highly glycosylated N-terminal ectodomain and a C-terminal hydrophobic anchor. After being synthesized the E1 glycoproteins associates with the E2 glycoprotein as a noncovalent heterodimer.
A model lipid bilayer is any bilayer assembled in vitro, as opposed to the bilayer of natural cell membranes or covering various sub-cellular structures like the nucleus. They are used to study the fundamental properties of biological membranes in a simplified and well-controlled environment, and increasingly in bottom-up synthetic biology for the construction of artificial cells. A model bilayer can be made with either synthetic or natural lipids. The simplest model systems contain only a single pure synthetic lipid. More physiologically relevant model bilayers can be made with mixtures of several synthetic or natural lipids.
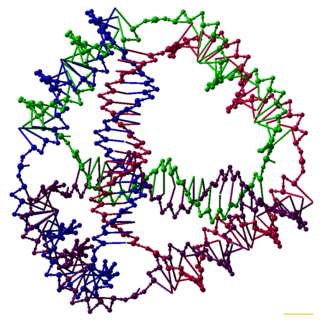
DNA nanotechnology is the design and manufacture of artificial nucleic acid structures for technological uses. In this field, nucleic acids are used as non-biological engineering materials for nanotechnology rather than as the carriers of genetic information in living cells. Researchers in the field have created static structures such as two- and three-dimensional crystal lattices, nanotubes, polyhedra, and arbitrary shapes, and functional devices such as molecular machines and DNA computers. The field is beginning to be used as a tool to solve basic science problems in structural biology and biophysics, including applications in X-ray crystallography and nuclear magnetic resonance spectroscopy of proteins to determine structures. Potential applications in molecular scale electronics and nanomedicine are also being investigated.

Bio-inspired photonics or bio-inspired optical materials are the application of biomimicry to the field of photonics. This differs slightly from biophotonics which is the study and manipulation of light to observe its interactions with biology. One area that inspiration may be drawn from is structural color, which allows color to appear as a result of the detailed material structure. Other inspiration can be drawn from both static and dynamic camouflage in animals like the chameleon or some cephalopods. Scientists have also been looking to recreate the ability to absorb light using molecules from various plants and microorganisms. Pulling from these heavily evolved constructs allows engineers to improve and optimize existing photonic technologies, whilst also solving existing problems within this field.
The SpyTag/SpyCatcher system is a technology for irreversible conjugation of recombinant proteins. The peptide SpyTag spontaneously reacts with the protein SpyCatcher to form an intermolecular isopeptide bond between the pair. DNA sequence encoding either SpyTag or SpyCatcher can be recombinantly introduced into the DNA sequence encoding a protein of interest, forming a fusion protein. These fusion proteins can be covalently linked when mixed in a reaction through the SpyTag/SpyCatcher system.













